The Draft Genome Sequence of the Yersinia entomophaga Entomopathogenic Type Strain MH96T
- PMID: 27187466
- PMCID: PMC4885058
- DOI: 10.3390/toxins8050143
The Draft Genome Sequence of the Yersinia entomophaga Entomopathogenic Type Strain MH96T
Abstract
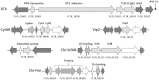
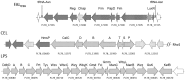
Here we report the draft genome of Yersinia entomophaga type strain MH96T. The genome shows 93.8% nucleotide sequence identity to that of Yersinia nurmii type strain APN3a-cT, and comprises a single chromosome of approximately 4,275,531 bp. In silico analysis identified that, in addition to the previously documented Y. entomophaga Yen-TC gene cluster, the genome encodes a diverse array of toxins, including two type III secretion systems, and five rhs-associated gene clusters. As well as these multicomponent systems, several orthologs of known insect toxins, such as VIP2 toxin and the binary toxin PirAB, and distant orthologs of some mammalian toxins, including repeats-in-toxin, a cytolethal distending toxin, hemolysin-like genes and an adenylate cyclase were identified. The genome also contains a large number of hypothetical proteins and orthologs of known effector proteins, such as LopT, as well as genes encoding a wide range of proteolytic determinants, including metalloproteases and pathogen fitness determinants, such as genes involved in iron metabolism. The bioinformatic data derived from the current in silico analysis, along with previous information on the pathobiology of Y. entomophaga against its insect hosts, suggests that a number of these virulence systems are required for survival in the hemocoel and incapacitation of the insect host.
Keywords: Rhs; Yersinia entomophaga; Yersinia nurmii; Yersinia ruckeri; entomopathogen; genome sequence.
Figures





Similar articles
-
Assessment of toxicity and persistence of Yersinia entomophaga and its Yen-Tc associated toxin.Pest Manag Sci. 2020 Dec;76(12):4301-4310. doi: 10.1002/ps.5997. Epub 2020 Aug 18. Pest Manag Sci. 2020. PMID: 32648630
-
Structural analysis of Chi1 Chitinase from Yen-Tc: the multisubunit insecticidal ABC toxin complex of Yersinia entomophaga.J Mol Biol. 2012 Jan 13;415(2):359-71. doi: 10.1016/j.jmb.2011.11.018. Epub 2011 Nov 15. J Mol Biol. 2012. PMID: 22108167
-
Temperature-Dependent Galleria mellonella Mortality as a Result of Yersinia entomophaga Infection.Appl Environ Microbiol. 2015 Sep;81(18):6404-14. doi: 10.1128/AEM.00790-15. Epub 2015 Jul 10. Appl Environ Microbiol. 2015. PMID: 26162867 Free PMC article.
-
Comparative genome analyses of the pathogenic Yersiniae based on the genome sequence of Yersinia enterocolitica strain 8081.Adv Exp Med Biol. 2007;603:2-16. doi: 10.1007/978-0-387-72124-8_1. Adv Exp Med Biol. 2007. PMID: 17966400 Review.
-
[The virulence genes of pathogenic Yersinia].Nihon Rinsho. 2003 Mar;61 Suppl 3:709-15. Nihon Rinsho. 2003. PMID: 12718052 Review. Japanese. No abstract available.
Cited by
-
Yersiniomics, a Multi-Omics Interactive Database for Yersinia Species.Microbiol Spectr. 2023 Feb 27;11(2):e0382622. doi: 10.1128/spectrum.03826-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36847572 Free PMC article.
-
Lysis Cassette-Mediated Exoprotein Release in Yersinia entomophaga Is Controlled by a PhoB-Like Regulator.Microbiol Spectr. 2023 Mar 23;11(2):e0036423. doi: 10.1128/spectrum.00364-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 36951587 Free PMC article.
-
Identification of genes involved in exoprotein release using a high-throughput exoproteome screening assay in Yersinia entomophaga.PLoS One. 2022 Jan 25;17(1):e0263019. doi: 10.1371/journal.pone.0263019. eCollection 2022. PLoS One. 2022. PMID: 35077520 Free PMC article.
-
Secretion Systems and Secreted Proteins in Gram-Negative Entomopathogenic Bacteria: Their Roles in Insect Virulence and Beyond.Insects. 2018 Jun 19;9(2):68. doi: 10.3390/insects9020068. Insects. 2018. PMID: 29921761 Free PMC article. Review.
-
A bacterial binary toxin system that kills both insects and aquatic crustaceans: Photorhabdus insect-related toxins A and B.PLoS Pathog. 2023 May 4;19(5):e1011330. doi: 10.1371/journal.ppat.1011330. eCollection 2023 May. PLoS Pathog. 2023. PMID: 37141203 Free PMC article. Review.
References
-
- Ewing W.H.A., Ross A.J., Brenner D.J., Fanning G.R. Yersinia ruckeri sp. nov., the redmouth (RM) bacterium. Int. J. Syst. Bacteriol. 1978;28:37–44. doi: 10.1099/00207713-28-1-37. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

