Mediator subunit Med12 contributes to the maintenance of neural stem cell identity
- PMID: 27188461
- PMCID: PMC4869265
- DOI: 10.1186/s12861-016-0114-0
Mediator subunit Med12 contributes to the maintenance of neural stem cell identity
Abstract
Background: The RNA polymerase II transcriptional Mediator subunit Med12 is broadly implicated in vertebrate brain development, and genetic variation in human MED12 is associated with X-linked intellectual disability and neuropsychiatric disorders. Although prior studies have begun to elaborate the functional contribution of Med12 within key neurodevelopmental pathways, a more complete description of Med12 function in the developing nervous system, including the specific biological networks and cellular processes under its regulatory influence, remains to be established. Herein, we sought to clarify the global contribution of Med12 to neural stem cell (NSC) biology through unbiased transcriptome profiling of mouse embryonic stem (ES) cell-derived NSCs following RNAi-mediated Med12 depletion.
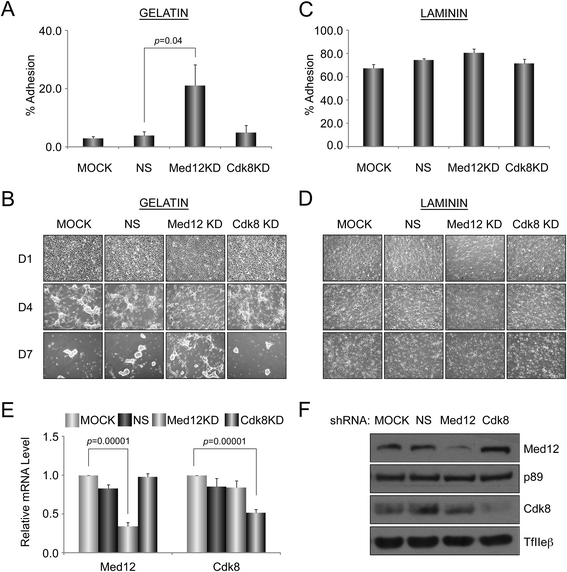
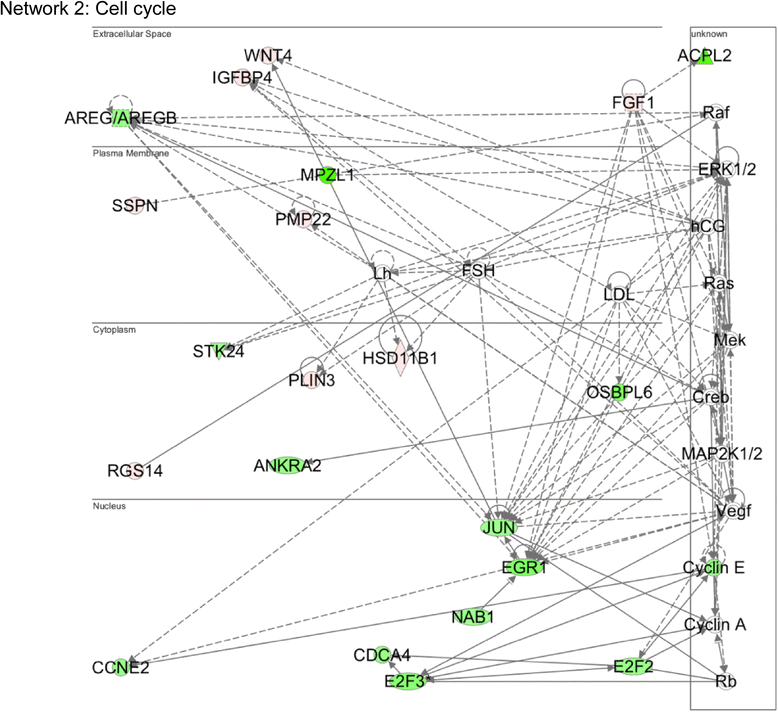
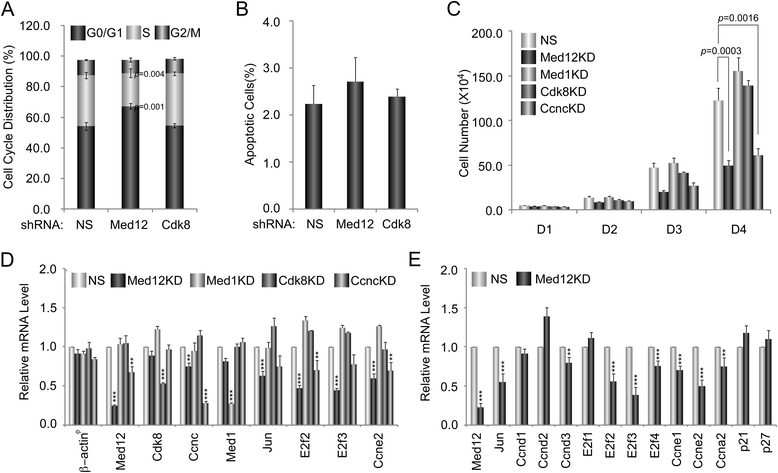
Results: A total of 240 genes (177 up, 73 down) were differentially expressed in Med12-knockdown versus control mouse NS-5 (mNS-5) NSCs. Gene set enrichment analysis revealed Med12 to be prominently linked with "cell-to-cell interaction" and "cell cycle" networks, and subsequent functional studies confirmed these associations. Targeted depletion of Med12 led to enhanced NSC adhesion and upregulation of cell adhesion genes, including Syndecan 2 (Sdc2). Concomitant depletion of both Sdc2 and Med12 reversed enhanced cell adhesion triggered by Med12 knockdown alone, confirming that Med12 negatively regulates NSC cell adhesion by suppressing the expression of cell adhesion molecules. Med12-mediated suppression of NSC adhesion is a dynamically regulated process in vitro, enforced in self-renewing NSCs and alleviated during the course of neuronal differentiation. Accordingly, Med12 depletion enhanced adhesion and prolonged survival of mNS-5 NSCs induced to differentiate on gelatin, effects that were bypassed completely by growth on laminin. On the other hand, Med12 depletion in mNS-5 NSCs led to reduced expression of G1/S phase cell cycle regulators and a concordant G1/S phase cell cycle block without evidence of apoptosis, resulting in a severe proliferation defect.
Conclusions: Med12 contributes to the maintenance of NSC identity through a functionally bipartite role in suppression and activation of gene expression programs dedicated to cell adhesion and G1/S phase cell cycle progression, respectively. Med12 may thus contribute to the regulatory apparatus that controls the balance between NSC self-renewal and differentiation, with important implications for MED12-linked neurodevelopmental disorders.
Keywords: Cell adhesion; Cell cycle; Gene expression; Med12; Mediator; Microarray; Neural stem cell.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

