Topological distortion and reorganized modular structure of gut microbial co-occurrence networks in inflammatory bowel disease
- PMID: 27188829
- PMCID: PMC4870640
- DOI: 10.1038/srep26087
Topological distortion and reorganized modular structure of gut microbial co-occurrence networks in inflammatory bowel disease
Abstract
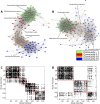
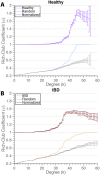
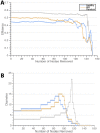
The gut microbiome plays a key role in human health, and alterations of the normal gut flora are associated with a variety of distinct disease states. Yet, the natural dependencies between microbes in healthy and diseased individuals remain far from understood. Here we use a network-based approach to characterize microbial co-occurrence in individuals with inflammatory bowel disease (IBD) and healthy (non-IBD control) individuals. We find that microbial networks in patients with IBD differ in both global structure and local connectivity patterns. While a "core" microbiome is preserved, network topology of other densely interconnected microbe modules is distorted, with potent inflammation-mediating organisms assuming roles as integrative and highly connected inter-modular hubs. We show that while both networks display a rich-club organization, in which a small set of microbes commonly co-occur, the healthy network is more easily disrupted by elimination of a small number of key species. Further investigation of network alterations in disease might offer mechanistic insights into the specific pathogens responsible for microbiome-mediated inflammation in IBD.
Figures


References
-
- Zhao L. The gut microbiota and obesity: from correlation to causality. Nature Rev Microbiol 11, 639–647 (2013). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

