Novel strong promoter of antimicrobial peptides gene pro-SmAMP2 from chickweed (Stellaria media)
- PMID: 27189173
- PMCID: PMC4870781
- DOI: 10.1186/s12896-016-0273-x
Novel strong promoter of antimicrobial peptides gene pro-SmAMP2 from chickweed (Stellaria media)
Abstract
Background: In a previous study we found that in chickweed the expression level of the pro-SmAMP2 gene was comparable or even higher to that of the β-actin gene. This high level of the gene expression has attracted our attention as an opportunity for the identification of novel strong promoters of plant origin, which could find its application in plant biotechnology. Therefore, in the present study we focused on the nucleotide sequence identification and the functional characteristics of the pro-SmAMP2 promoter in transgenic plants.
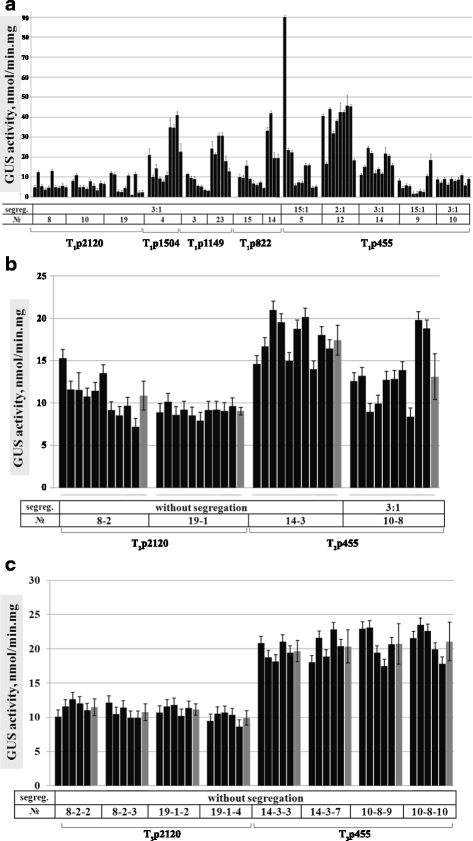
Results: In chickweed (Stellaria media), a 2120 bp promoter region of the pro-SmAMP2 gene encoding antifungal peptides was sequenced. Six 5'-deletion variants -2120, -1504, -1149, -822, -455, and -290 bp of pro-SmAMP2 gene promoter were fused with the coding region of the reporter gene gusA in the plant expression vector pCambia1381Z. Independent transgenic plants of tobacco Nicotiana tabacum were obtained with each genetic structure. GUS protein activity assay in extracts from transgenic plants showed that all deletion variants of the promoter, except -290 bp, expressed the gusA gene. In most transgenic plants, the GUS activity level was comparable or higher than in plants with the viral promoter CaMV 35S. GUS activity remains high in progenies and its level correlates positively with the amount of gusA gene mRNA in T3 homozygous plants. The activity of the рro-SmAMP2 promoter was detected in all organs of the transgenic plants studied, during meiosis and in pollen as well.
Conclusion: Our results show that the рro-SmAMP2 promoter can be used for target genes expression control in transgenic plants.
Keywords: Expression control; Nicotiana tabacum; Promoter; Stellaria media; Transgenic plants; pro-SmAMP2.
Figures







References
-
- Potenza C, Aleman L, Sengupta-Gopalan C. Targeting transgene expression in research, agricultural, and environmental applications: Promoters used in plant transformation. In Vitro Cell Dev Biol-Plant. 2004;40(1):1–22. doi: 10.1079/IVP2003477. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

