Conservation and Innovation of APOBEC3A Restriction Functions during Primate Evolution
- PMID: 27189538
- PMCID: PMC5010000
- DOI: 10.1093/molbev/msw070
Conservation and Innovation of APOBEC3A Restriction Functions during Primate Evolution
Abstract
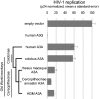
LINE-1 (long interspersed element-1) retroelements are the only active autonomous endogenous retroelements in human genomes. Their retrotransposition activity has created close to 50% of the current human genome. Due to the apparent costs of this proliferation, host genomes have evolved multiple mechanisms to curb LINE-1 retrotransposition. Here, we investigate the evolution and function of the LINE-1 restriction factor APOBEC3A, a member of the APOBEC3 cytidine deaminase gene family. We find that APOBEC3A genes have evolved rapidly under diversifying selection in primates, suggesting changes in APOBEC3A have been recurrently selected in a host-pathogen "arms race." Nonetheless, in contrast to previous reports, we find that the LINE-1 restriction activity of APOBEC3A proteins has been strictly conserved throughout simian primate evolution in spite of its pervasive diversifying selection. Based on these results, we conclude that LINE-1s have not driven the rapid evolution of APOBEC3A in primates. In contrast to this conserved LINE-1 restriction, we find that a subset of primate APOBEC3A genes have enhanced antiviral restriction. We trace this gain of antiviral restriction in APOBEC3A to the common ancestor of a subset of Old World monkeys. Thus, APOBEC3A has not only maintained its LINE-1 restriction ability, but also evolved a gain of antiviral specificity against other pathogens. Our findings suggest that while APOBEC3A has evolved to restrict additional pathogens, only those adaptive amino acid changes that leave LINE-1 restriction unperturbed have been tolerated.
Keywords: APOBEC3A; LINE-1; innate immunity; positive selection; primates.; restriction factor; retroelements.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



References
-
- Aguiar RS, Lovsin N, Tanuri A, Peterlin BM. 2008. Vpr.A3A chimera inhibits HIV replication. J Biol Chem. 283:2518–2525. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

