Pooled Enrichment Sequencing Identifies Diversity and Evolutionary Pressures at NLR Resistance Genes within a Wild Tomato Population
- PMID: 27189991
- PMCID: PMC4898808
- DOI: 10.1093/gbe/evw094
Pooled Enrichment Sequencing Identifies Diversity and Evolutionary Pressures at NLR Resistance Genes within a Wild Tomato Population
Abstract
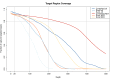
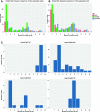
Nod-like receptors (NLRs) are nucleotide-binding domain and leucine-rich repeats containing proteins that are important in plant resistance signaling. Many of the known pathogen resistance (R) genes in plants are NLRs and they can recognize pathogen molecules directly or indirectly. As such, divergence and copy number variants at these genes are found to be high between species. Within populations, positive and balancing selection are to be expected if plants coevolve with their pathogens. In order to understand the complexity of R-gene coevolution in wild nonmodel species, it is necessary to identify the full range of NLRs and infer their evolutionary history. Here we investigate and reveal polymorphism occurring at 220 NLR genes within one population of the partially selfing wild tomato species Solanum pennellii. We use a combination of enrichment sequencing and pooling ten individuals, to specifically sequence NLR genes in a resource and cost-effective manner. We focus on the effects which different mapping and single nucleotide polymorphism calling software and settings have on calling polymorphisms in customized pooled samples. Our results are accurately verified using Sanger sequencing of polymorphic gene fragments. Our results indicate that some NLRs, namely 13 out of 220, have maintained polymorphism within our S. pennellii population. These genes show a wide range of πN/πS ratios and differing site frequency spectra. We compare our observed rate of heterozygosity with expectations for this selfing and bottlenecked population. We conclude that our method enables us to pinpoint NLR genes which have experienced natural selection in their habitat.
Keywords: RENSeq; Solanum penellii; population genetics; resistance genes.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Evolution of NLR resistance genes with noncanonical N-terminal domains in wild tomato species.New Phytol. 2020 Sep;227(5):1530-1543. doi: 10.1111/nph.16628. Epub 2020 May 23. New Phytol. 2020. PMID: 32344448
-
Genome-wide identification of the NLR gene family in Haynaldia villosa by SMRT-RenSeq.BMC Genomics. 2022 Feb 10;23(1):118. doi: 10.1186/s12864-022-08334-w. BMC Genomics. 2022. PMID: 35144544 Free PMC article.
-
Subsets of NLR genes show differential signatures of adaptation during colonization of new habitats.New Phytol. 2019 Oct;224(1):367-379. doi: 10.1111/nph.16017. Epub 2019 Aug 8. New Phytol. 2019. PMID: 31230368
-
Evolution of Plant NLRs: From Natural History to Precise Modifications.Annu Rev Plant Biol. 2020 Apr 29;71:355-378. doi: 10.1146/annurev-arplant-081519-035901. Epub 2020 Feb 24. Annu Rev Plant Biol. 2020. PMID: 32092278 Review.
-
The Tomato Interspecific NB-LRR Gene Arsenal and Its Impact on Breeding Strategies.Genes (Basel). 2021 Jan 27;12(2):184. doi: 10.3390/genes12020184. Genes (Basel). 2021. PMID: 33514027 Free PMC article. Review.
Cited by
-
Genome assembly and characterization of a complex zfBED-NLR gene-containing disease resistance locus in Carolina Gold Select rice with Nanopore sequencing.PLoS Genet. 2020 Jan 27;16(1):e1008571. doi: 10.1371/journal.pgen.1008571. eCollection 2020 Jan. PLoS Genet. 2020. PMID: 31986137 Free PMC article.
-
Evolution of pathogen response genes associated with increased disease susceptibility during adaptation to an extreme drought in a Brassica rapa plant population.BMC Ecol Evol. 2021 Apr 21;21(1):61. doi: 10.1186/s12862-021-01789-7. BMC Ecol Evol. 2021. PMID: 33882815 Free PMC article.
-
The de Novo Reference Genome and Transcriptome Assemblies of the Wild Tomato Species Solanum chilense Highlights Birth and Death of NLR Genes Between Tomato Species.G3 (Bethesda). 2019 Dec 3;9(12):3933-3941. doi: 10.1534/g3.119.400529. G3 (Bethesda). 2019. PMID: 31604826 Free PMC article.
-
Genomic structural variation in tomato and its role in plant immunity.Mol Hortic. 2022 Mar 10;2(1):7. doi: 10.1186/s43897-022-00029-w. Mol Hortic. 2022. PMID: 37789472 Free PMC article. Review.
-
Parallel evolution of the POQR prolyl oligo peptidase gene conferring plant quantitative disease resistance.PLoS Genet. 2017 Dec 22;13(12):e1007143. doi: 10.1371/journal.pgen.1007143. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29272270 Free PMC article.
References
-
- 100 Tomato Genome Sequencing Consortium, et al. 2014. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 80:136–148. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

