C-Terminal Region of Sulfite Reductase Is Important to Localize to Chloroplast Nucleoids in Land Plants
- PMID: 27189994
- PMCID: PMC4898807
- DOI: 10.1093/gbe/evw093
C-Terminal Region of Sulfite Reductase Is Important to Localize to Chloroplast Nucleoids in Land Plants
Abstract
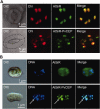
Chloroplast (cp) DNA is compacted into cpDNA-protein complexes, called cp nucleoids. An abundant and extensively studied component of cp nucleoids is the bifunctional protein sulfite reductase (SiR). The preconceived role of SiR as the core cp nucleoid protein, however, is becoming less likely because of the recent findings that SiRs do not associate with cp nucleoids in some plant species, such as Zea mays and Arabidopsis thaliana To address this discrepancy, we have performed a detailed phylogenetic analysis of SiRs, which shows that cp nucleoid-type SiRs share conserved C-terminally encoded peptides (CEPs). The CEPs are likely to form a bacterial ribbon-helix-helix DNA-binding motif, implying a potential role in attaching SiRs onto cp nucleoids. A proof-of-concept experiment was conducted by fusing the nonnucleoid-type SiR from A. thaliana (AtSiR) with the CEP from the cp nucleoid-type SiR of Phaseolus vulgaris The addition of the CEP drastically altered the intra-cp localization of AtSiR to cp nucleoids. Our analysis supports the possible functions of CEPs in determining the localization of SiRs to cp nucleoids and illuminates a possible evolutionary scenario for SiR as a cp nucleoid protein.
Keywords: evolution; nucleoid; sulfite reductase.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

