Capturing the Phylogeny of Holometabola with Mitochondrial Genome Data and Bayesian Site-Heterogeneous Mixture Models
- PMID: 27189999
- PMCID: PMC4898802
- DOI: 10.1093/gbe/evw086
Capturing the Phylogeny of Holometabola with Mitochondrial Genome Data and Bayesian Site-Heterogeneous Mixture Models
Abstract
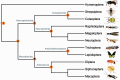
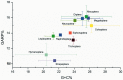
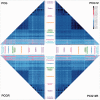
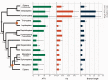
After decades of debate, a mostly satisfactory resolution of relationships among the 11 recognized holometabolan orders of insects has been reached based on nuclear genes, resolving one of the most substantial branches of the tree-of-life, but the relationships are still not well established with mitochondrial genome data. The main reasons have been the absence of sufficient data in several orders and lack of appropriate phylogenetic methods that avoid the systematic errors from compositional and mutational biases in insect mitochondrial genomes. In this study, we assembled the richest taxon sampling of Holometabola to date (199 species in 11 orders), and analyzed both nucleotide and amino acid data sets using several methods. We find the standard Bayesian inference and maximum-likelihood analyses were strongly affected by systematic biases, but the site-heterogeneous mixture model implemented in PhyloBayes avoided the false grouping of unrelated taxa exhibiting similar base composition and accelerated evolutionary rate. The inclusion of rRNA genes and removal of fast-evolving sites with the observed variability sorting method for identifying sites deviating from the mean rates improved the phylogenetic inferences under a site-heterogeneous model, correctly recovering most deep branches of the Holometabola phylogeny. We suggest that the use of mitochondrial genome data for resolving deep phylogenetic relationships requires an assessment of the potential impact of substitutional saturation and compositional biases through data deletion strategies and by using site-heterogeneous mixture models. Our study suggests a practical approach for how to use densely sampled mitochondrial genome data in phylogenetic analyses.
Keywords: Holometabola phylogeny; PhyloBayes; compositional bias; mitochondrial phylogenomics; rate variation; tree-of-life.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

Similar articles
-
What is the phylogenetic signal limit from mitogenomes? The reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny.BMC Evol Biol. 2011 Oct 27;11:315. doi: 10.1186/1471-2148-11-315. BMC Evol Biol. 2011. PMID: 22032248 Free PMC article.
-
Compositional and mutational rate heterogeneity in mitochondrial genomes and its effect on the phylogenetic inferences of Cimicomorpha (Hemiptera: Heteroptera).BMC Genomics. 2018 Apr 18;19(1):264. doi: 10.1186/s12864-018-4650-9. BMC Genomics. 2018. PMID: 29669515 Free PMC article.
-
Mitochondrial phylogenomics of early land plants: mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias.Syst Biol. 2014 Nov;63(6):862-78. doi: 10.1093/sysbio/syu049. Epub 2014 Jul 28. Syst Biol. 2014. PMID: 25070972
-
A comparative analysis of complete mitochondrial genomes among Hexapoda.Mol Phylogenet Evol. 2013 Nov;69(2):393-403. doi: 10.1016/j.ympev.2013.03.033. Epub 2013 Apr 15. Mol Phylogenet Evol. 2013. PMID: 23598069 Review.
-
Insect mitochondrial genomics: implications for evolution and phylogeny.Annu Rev Entomol. 2014;59:95-117. doi: 10.1146/annurev-ento-011613-162007. Epub 2013 Oct 16. Annu Rev Entomol. 2014. PMID: 24160435 Review.
Cited by
-
The first mitogenome of the subfamily Stenoponiinae (Siphonaptera: Ctenophthalmidae) and implications for its phylogenetic position.Sci Rep. 2024 Aug 6;14(1):18179. doi: 10.1038/s41598-024-69203-y. Sci Rep. 2024. PMID: 39107455 Free PMC article.
-
Mitochondrial genomes provide insights into the Euholognatha (Insecta: Plecoptera).BMC Ecol Evol. 2024 Feb 1;24(1):16. doi: 10.1186/s12862-024-02205-6. BMC Ecol Evol. 2024. PMID: 38297210 Free PMC article.
-
A Mitochondrial Genome Phylogeny of Cleridae (Coleoptera, Cleroidea).Insects. 2022 Jan 24;13(2):118. doi: 10.3390/insects13020118. Insects. 2022. PMID: 35206692 Free PMC article.
-
Mitochondrial genomes of the stoneflies Mesonemourametafiligera and Mesonemouratritaenia (Plecoptera, Nemouridae), with a phylogenetic analysis of Nemouroidea.Zookeys. 2019 Apr 4;835:43-63. doi: 10.3897/zookeys.835.32470. eCollection 2019. Zookeys. 2019. PMID: 31043849 Free PMC article.
-
A Comparative Analysis and Limited Phylogenetic Implications of Mitogenomes in Infraorder-Level Diptera.Int J Mol Sci. 2025 Jul 25;26(15):7222. doi: 10.3390/ijms26157222. Int J Mol Sci. 2025. PMID: 40806355 Free PMC article.
References
-
- Abascal F, Posada D, Zardoya R. 2007. MtArt: a new model of amino acid replacement for Arthropoda. Mol Biol Evol. 24:1–5. - PubMed
-
- Baurain D, Brinkmann H, Philippe H. 2007. Lack of resolution in the animal phylogeny: closely spaced cladogeneses or undetected systematic errors? Mol Biol Evol. 24:6–9. - PubMed
-
- Bergsten J. 2005. A review of long-branch attraction. Cladistics 21:163–193. - PubMed
-
- Bernt M, et al. 2013. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol Phylogenet Evol. 69:252–364. - PubMed
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

