Incoming human papillomavirus type 16 genome resides in a vesicular compartment throughout mitosis
- PMID: 27190090
- PMCID: PMC4896702
- DOI: 10.1073/pnas.1600638113
Incoming human papillomavirus type 16 genome resides in a vesicular compartment throughout mitosis
Abstract
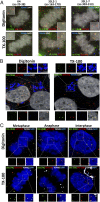
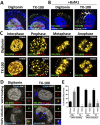
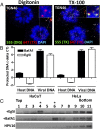
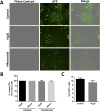
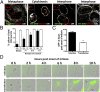
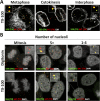
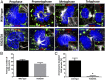
During the entry process, the human papillomavirus (HPV) capsid is trafficked to the trans-Golgi network (TGN), whereupon it enters the nucleus during mitosis. We previously demonstrated that the minor capsid protein L2 assumes a transmembranous conformation in the TGN. Here we provide evidence that the incoming viral genome dissociates from the TGN and associates with microtubules after the onset of mitosis. Deposition onto mitotic chromosomes is L2-mediated. Using differential staining of an incoming viral genome by small molecular dyes in selectively permeabilized cells, nuclease protection, and flotation assays, we found that HPV resides in a membrane-bound vesicle until mitosis is completed and the nuclear envelope has reformed. As a result, expression of the incoming viral genome is delayed. Taken together, these data provide evidence that HPV has evolved a unique strategy for delivering the viral genome to the nucleus of dividing cells. Furthermore, it is unlikely that nuclear vesicles are unique to HPV, and thus we may have uncovered a hitherto unrecognized cellular pathway that may be of interest for future cell biological studies.
Keywords: HPV entry; digitonin; mitosis; nuclear vesicle; vesicular transport.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
HPV16 entry requires dynein for minus-end transport and utilizes kinesin Kif11 for plus-end transport along microtubules during mitosis.J Virol. 2025 Jan 31;99(1):e0093724. doi: 10.1128/jvi.00937-24. Epub 2024 Dec 4. J Virol. 2025. PMID: 39629998 Free PMC article.
-
Translocation of the papillomavirus L2/vDNA complex across the limiting membrane requires the onset of mitosis.PLoS Pathog. 2017 May 2;13(5):e1006200. doi: 10.1371/journal.ppat.1006200. eCollection 2017 May. PLoS Pathog. 2017. PMID: 28463988 Free PMC article.
-
Human Papillomavirus 16 Infection Induces VAP-Dependent Endosomal Tubulation.J Virol. 2018 Feb 26;92(6):e01514-17. doi: 10.1128/JVI.01514-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29321327 Free PMC article.
-
Human Papillomavirus Entry: Hiding in a Bubble.J Virol. 2016 Aug 26;90(18):8032-5. doi: 10.1128/JVI.01065-16. Print 2016 Sep 15. J Virol. 2016. PMID: 27412595 Free PMC article. Review.
-
Cruising the cellular highways: How human papillomavirus travels from the surface to the nucleus.Virus Res. 2017 Mar 2;231:1-9. doi: 10.1016/j.virusres.2016.10.015. Epub 2016 Oct 29. Virus Res. 2017. PMID: 27984059 Free PMC article. Review.
Cited by
-
Synonymous nucleotide changes drive papillomavirus evolution.Tumour Virus Res. 2022 Dec;14:200248. doi: 10.1016/j.tvr.2022.200248. Epub 2022 Oct 17. Tumour Virus Res. 2022. PMID: 36265836 Free PMC article. Review.
-
Non-enveloped virus membrane penetration: New advances leading to new insights.PLoS Pathog. 2022 Dec 8;18(12):e1010948. doi: 10.1371/journal.ppat.1010948. eCollection 2022 Dec. PLoS Pathog. 2022. PMID: 36480535 Free PMC article.
-
Master mitotic kinases regulate viral genome delivery during papillomavirus cell entry.Nat Commun. 2023 Jan 23;14(1):355. doi: 10.1038/s41467-023-35874-w. Nat Commun. 2023. PMID: 36683055 Free PMC article.
-
α-Defensin HD5 Inhibits Human Papillomavirus 16 Infection via Capsid Stabilization and Redirection to the Lysosome.mBio. 2017 Jan 24;8(1):e02304-16. doi: 10.1128/mBio.02304-16. mBio. 2017. PMID: 28119475 Free PMC article.
-
Activating the DNA Damage Response and Suppressing Innate Immunity: Human Papillomaviruses Walk the Line.Pathogens. 2020 Jun 13;9(6):467. doi: 10.3390/pathogens9060467. Pathogens. 2020. PMID: 32545729 Free PMC article. Review.
References
-
- Buck CB, Pastrana DV, Lowy DR, Schiller JT. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med. 2005;119:445–462. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

