Parallel Evolution in Streptococcus pneumoniae Biofilms
- PMID: 27190203
- PMCID: PMC4898793
- DOI: 10.1093/gbe/evw072
Parallel Evolution in Streptococcus pneumoniae Biofilms
Abstract
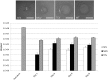
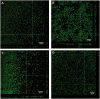
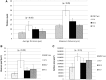
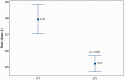
Streptococcus pneumoniae is a commensal human pathogen and the causative agent of various invasive and noninvasive diseases. Carriage of the pneumococcus in the nasopharynx is thought to be mediated by biofilm formation, an environment where isogenic populations frequently give rise to morphological colony variants, including small colony variant (SCV) phenotypes. We employed metabolic characterization and whole-genome sequencing of biofilm-derived S. pneumoniae serotype 22F pneumococcal SCVs to investigate diversification during biofilm formation. Phenotypic profiling revealed that SCVs exhibit reduced growth rates, reduced capsule expression, altered metabolic profiles, and increased biofilm formation compared to the ancestral strain. Whole-genome sequencing of 12 SCVs from independent biofilm experiments revealed that all SCVs studied had mutations within the DNA-directed RNA polymerase delta subunit (RpoE). Mutations included four large-scale deletions ranging from 51 to 264 bp, one insertion resulting in a coding frameshift, and seven nonsense single-nucleotide substitutions that result in a truncated gene product. This work links mutations in the rpoE gene to SCV formation and enhanced biofilm development in S. pneumoniae and therefore may have important implications for colonization, carriage, and persistence of the organism. Furthermore, recurrent mutation of the pneumococcal rpoE gene presents an unprecedented level of parallel evolution in pneumococcal biofilm development.
Keywords: Streptococcus pneumoniae; biofilm; genomic diversification; parallel evolution.
© The Author(s) 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Achberger EC, Whiteley HR. 1981. The role of the delta-peptide of the Bacillus-subtilis RNA-polymerase in promoter selection. J Biol Chem. 256:7424–7432. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

