The Occurrence of Genetic Alterations during the Progression of Breast Carcinoma
- PMID: 27190992
- PMCID: PMC4848409
- DOI: 10.1155/2016/5237827
The Occurrence of Genetic Alterations during the Progression of Breast Carcinoma
Abstract
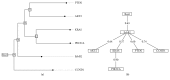
The interrelationship among genetic variations between the developing process of carcinoma and the order of occurrence has not been completely understood. Interpreting the mechanisms of copy number variation (CNV) is absolutely necessary for understanding the etiology of genetic disorders. Oncogenetic tree is a special phylogenetic tree inferential pictorial representation of oncogenesis. In our present study, we constructed oncogenetic tree to imitate the occurrence of genetic and cytogenetic alterations in human breast cancer. The oncogenetic tree model was built on CNV of ErbB2, AKT2, KRAS, PIK3CA, PTEN, and CCND1 genes in 963 cases of tumors with sequencing and CNA data of human breast cancer from TCGA. Results from the oncogenetic tree model indicate that ErbB2 copy number variation is the frequent early event of human breast cancer. The oncogenetic tree model based on the phylogenetic tree is a type of mathematical model that may eventually provide a better way to understand the process of oncogenesis.
Figures


Similar articles
-
Somatic mutations in epidermal growth factor receptor signaling pathway genes in non-small cell lung cancers.J Thorac Oncol. 2010 Nov;5(11):1734-40. doi: 10.1097/JTO.0b013e3181f0beca. J Thorac Oncol. 2010. PMID: 20881644
-
CCND1- and ERBB2-gene deregulation and PTEN mutation analyses in invasive lobular carcinoma of the breast.Mol Carcinog. 2002 Sep;35(1):6-12. doi: 10.1002/mc.10069. Mol Carcinog. 2002. PMID: 12203362
-
Evaluation of the association of PIK3CA mutations and PTEN loss with efficacy of trastuzumab therapy in metastatic breast cancer.Breast Cancer Res Treat. 2011 Jul;128(2):447-56. doi: 10.1007/s10549-011-1572-5. Epub 2011 May 19. Breast Cancer Res Treat. 2011. PMID: 21594665
-
Correlation between activation of PI3K/AKT/mTOR pathway and prognosis of breast cancer in Chinese women.PLoS One. 2015 Mar 27;10(3):e0120511. doi: 10.1371/journal.pone.0120511. eCollection 2015. PLoS One. 2015. PMID: 25816324 Free PMC article.
-
Highly prevalent genetic alterations in receptor tyrosine kinases and phosphatidylinositol 3-kinase/akt and mitogen-activated protein kinase pathways in anaplastic and follicular thyroid cancers.J Clin Endocrinol Metab. 2008 Aug;93(8):3106-16. doi: 10.1210/jc.2008-0273. Epub 2008 May 20. J Clin Endocrinol Metab. 2008. PMID: 18492751
Cited by
-
Smoking alters the evolutionary trajectory of non-small cell lung cancer.Exp Ther Med. 2019 Nov;18(5):3315-3324. doi: 10.3892/etm.2019.7958. Epub 2019 Aug 29. Exp Ther Med. 2019. PMID: 31602204 Free PMC article.
-
Identifying Methylation Pattern and Genes Associated with Breast Cancer Subtypes.Int J Mol Sci. 2019 Aug 31;20(17):4269. doi: 10.3390/ijms20174269. Int J Mol Sci. 2019. PMID: 31480430 Free PMC article.
-
Multiclass Cancer Prediction Based on Copy Number Variation Using Deep Learning.Comput Intell Neurosci. 2022 Jun 9;2022:4742986. doi: 10.1155/2022/4742986. eCollection 2022. Comput Intell Neurosci. 2022. PMID: 35720914 Free PMC article.
-
Copy number variation heterogeneity reveals biological inconsistency in hierarchical cancer classifications.Mol Cytogenet. 2024 Nov 6;17(1):26. doi: 10.1186/s13039-024-00692-2. Mol Cytogenet. 2024. PMID: 39506842 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

