Notch1 pathway-mediated microRNA-151-5p promotes gastric cancer progression
- PMID: 27191259
- PMCID: PMC5122370
- DOI: 10.18632/oncotarget.9342
Notch1 pathway-mediated microRNA-151-5p promotes gastric cancer progression
Abstract
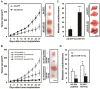
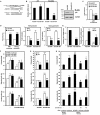
Gastric carcinoma is the third leading cause of lethal cancer worldwide. Previous studies showed that Notch1 receptor intracellular domain (N1IC), the activated form of Notch1 receptor, promotes gastric cancer progression. It has been demonstrated that a significant cross-talk interplays between Notch pathways and microRNAs (miRNAs) in controlling tumorigenesis. This study identified an intronic microRNA-151 (miR-151), which consists of two mature miRNAs, miR-151-3p and miR-151-5p, as a Notch1 receptor-induced miRNA in gastric cancer cells. Activation of Notch1 pathway enhanced expressions of miR-151 and its host gene, focal adhesion kinase (FAK), in gastric cancer cells. The levels of miR-151 in gastric cancer samples were higher than those of adjacent non-tumor samples. Activated Notch1 pathway induced CBF1-dependent FAK promoter activity. The ectopic expression of miR-151 promoted growth and progression of SC-M1 gastric cancer cells including cell viability and colony formation, migration, and invasion abilities. Activated Notch1 pathway could augment progression of gastric cancer cells through miR-151-5p and FAK. The mRNA levels of pluripotency genes, Nanog and SOX-2, tumorsphere formation ability, tumor growth, and lung metastasis of SC-M1 cells were elevated by activated Notch1 pathway through miR-151-5p. Furthermore, miR-151-5p could target 3'-untranslated region (3'-UTR) of p53 mRNA and down-regulate p53 level in SC-M1 cells. Mechanistically, Notch1/miR-151-5p axis contributed to progression of SC-M1 cells through down-regulation of p53 which in turn repressed FAK promoter activity. Taken together, these results suggest that Notch1 pathway and miR-151-5p interplay with p53 in a reciprocal regulation loop in controlling gastric carcinogenesis.
Keywords: FAK; Notch1 receptor; gastric carcinogenesis; miR-151; p53.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures






References
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. - PubMed
-
- Yin L, Velazquez OC, Liu ZJ. Notch signaling: emerging molecular targets for cancer therapy. Biochem Pharmacol. 2010;80:690–701. - PubMed
-
- Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling--are we there yet? Nat Rev Drug Discov. 2014;13:357–378. - PubMed
-
- Tseng YC, Tsai YH, Tseng MJ, Hsu KW, Yang MC, Huang KH, Li AF, Chi CW, Hsieh RH, Ku HH, Yeh TS. Notch2-induced COX-2 expression enhancing gastric cancer progression. Mol Carcinog. 2012;51:939–951. - PubMed
-
- Yeh TS, Wu CW, Hsu KW, Liao WJ, Yang MC, Li AF, Wang AM, Kuo ML, Chi CW. The activated Notch1 signal pathway is associated with gastric cancer progression through cyclooxygenase-2. Cancer Res. 2009;69:5039–5048. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

