Deep biomarkers of human aging: Application of deep neural networks to biomarker development
- PMID: 27191382
- PMCID: PMC4931851
- DOI: 10.18632/aging.100968
Deep biomarkers of human aging: Application of deep neural networks to biomarker development
Abstract
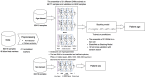
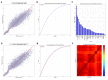
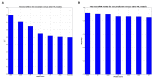
One of the major impediments in human aging research is the absence of a comprehensive and actionable set of biomarkers that may be targeted and measured to track the effectiveness of therapeutic interventions. In this study, we designed a modular ensemble of 21 deep neural networks (DNNs) of varying depth, structure and optimization to predict human chronological age using a basic blood test. To train the DNNs, we used over 60,000 samples from common blood biochemistry and cell count tests from routine health exams performed by a single laboratory and linked to chronological age and sex. The best performing DNN in the ensemble demonstrated 81.5 % epsilon-accuracy r = 0.90 with R(2) = 0.80 and MAE = 6.07 years in predicting chronological age within a 10 year frame, while the entire ensemble achieved 83.5% epsilon-accuracy r = 0.91 with R(2) = 0.82 and MAE = 5.55 years. The ensemble also identified the 5 most important markers for predicting human chronological age: albumin, glucose, alkaline phosphatase, urea and erythrocytes. To allow for public testing and evaluate real-life performance of the predictor, we developed an online system available at http://www.aging.ai. The ensemble approach may facilitate integration of multi-modal data linked to chronological age and sex that may lead to simple, minimally invasive, and affordable methods of tracking integrated biomarkers of aging in humans and performing cross-species feature importance analysis.
Keywords: aging biomarkers; biomarker development; deep learning; deep neural networks; human aging; machine learning.
Conflict of interest statement
The authors are affiliated with Insilico Medicine, Inc, a commercial company developing differential pathway activation scoring-based and deep learned biomarkers of multiple diseases and aging and engaging in drug discovery and drug repurposing. The company has developed a range of drug candidates addressing specific diseases and geroprotector interventions addressing human aging processes that need to be validated in human patients. The company intends to use blood biochemistry and multi-parametric markers, including the one published in this paper to test the efficacy of these compounds. Despite company's commitment to best academic practices and in silico veritas, the authors may have a conflict of interest.
Figures


Comment in
-
Deep biomarkers of aging are population-dependent.Aging (Albany NY). 2016 Sep 8;8(9):2253-2255. doi: 10.18632/aging.101034. Aging (Albany NY). 2016. PMID: 27622833 Free PMC article. No abstract available.
References
-
- Moskalev A, Chernyagina E, de Magalhães JP, Barardo D, Thoppil H, Shaposhnikov M, et al. Geroprotectors.org: a new, structured and curated database of current therapeutic interventions in aging and age-related disease. Aging (Albany NY) 2015;7:616–728. - PMC - PubMed
-
- Bürkle A, Moreno-Villanueva M, Bernhard J, Blasco M, Zondag G, Hoeijmakers JHJ, et al. MARK-AGE biomarkers of ageing. Mech. Ageing Dev. 2015;151:2–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

