Variable presence of the inverted repeat and plastome stability in Erodium
- PMID: 27192713
- PMCID: PMC4904181
- DOI: 10.1093/aob/mcw065
Variable presence of the inverted repeat and plastome stability in Erodium
Abstract
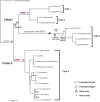
Background and aims: Several unrelated lineages such as plastids, viruses and plasmids, have converged on quadripartite genomes of similar size with large and small single copy regions and a large inverted repeat (IR). Except for Erodium (Geraniaceae), saguaro cactus and some legumes, the plastomes of all photosynthetic angiosperms display this structure. The functional significance of the IR is not understood and Erodium provides a system to examine the role of the IR in the long-term stability of these genomes. We compared the degree of genomic rearrangement in plastomes of Erodium that differ in the presence and absence of the IR.
Methods: We sequenced 17 new Erodium plastomes. Using 454, Illumina, PacBio and Sanger sequences, 16 genomes were assembled and categorized along with one incomplete and two previously published Erodium plastomes. We conducted phylogenetic analyses among these species using a dataset of 19 protein-coding genes and determined if significantly higher evolutionary rates had caused the long branch seen previously in phylogenetic reconstructions within the genus. Bioinformatic comparisons were also performed to evaluate plastome evolution across the genus.
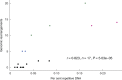
Key results: Erodium plastomes fell into four types (Type 1-4) that differ in their substitution rates, short dispersed repeat content and degree of genomic rearrangement, gene and intron content and GC content. Type 4 plastomes had significantly higher rates of synonymous substitutions (dS) for all genes and for 14 of the 19 genes non-synonymous substitutions (dN) were significantly accelerated. We evaluated the evidence for a single IR loss in Erodium and in doing so discovered that Type 4 plastomes contain a novel IR.
Conclusions: The presence or absence of the IR does not affect plastome stability in Erodium. Rather, the overall repeat content shows a negative correlation with genome stability, a pattern in agreement with other angiosperm groups and recent findings on genome stability in bacterial endosymbionts.
Keywords: Erodium; Geraniaceae; chloroplast; genome evolution; inversions; inverted repeat; plastome; repeated sequences.
© The Author 2016. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Blazier JC, Guisinger MM, Jansen RK. 2011. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Molecular Biology 76: 263–272. - PubMed
-
- Bock R. 2007. Structure, function, and inheritance of plastomes In: Bock R, ed. Cell and molecular biology of plastids. Topics in current genetics. Berlin: Springer, 29–63.
-
- Broach JR, Volkert FC. 1991. Circular DNA plasmids of yeasts In: Broach JR, Pringle JR, Jones EW, eds. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics, Vol. I New York: Cold Spring Harbor Laboratory Press, 297–331.
-
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. In: Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB), 45–56. doi:10.1.1.23.7465.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

