A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation
- PMID: 27193062
- PMCID: PMC4874031
- DOI: 10.1038/ncomms11616
A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation
Abstract
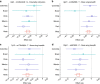
We report a genome-wide association scan for facial features in ∼6,000 Latin Americans. We evaluated 14 traits on an ordinal scale and found significant association (P values<5 × 10(-8)) at single-nucleotide polymorphisms (SNPs) in four genomic regions for three nose-related traits: columella inclination (4q31), nose bridge breadth (6p21) and nose wing breadth (7p13 and 20p11). In a subsample of ∼3,000 individuals we obtained quantitative traits related to 9 of the ordinal phenotypes and, also, a measure of nasion position. Quantitative analyses confirmed the ordinal-based associations, identified SNPs in 2q12 associated to chin protrusion, and replicated the reported association of nasion position with SNPs in PAX3. Strongest association in 2q12, 4q31, 6p21 and 7p13 was observed for SNPs in the EDAR, DCHS2, RUNX2 and GLI3 genes, respectively. Associated SNPs in 20p11 extend to PAX1. Consistent with the effect of EDAR on chin protrusion, we documented alterations of mandible length in mice with modified Edar funtion.
Figures



References
-
- Reyes-Centeno H., Hubbe M., Hanihara T., Stringer C. & Harvati K. Testing modern human out-of-Africa dispersal models and implications for modern human origins. J. Hum. Evol. 87, 95–106 (2015). - PubMed
-
- Harvati K. & Weaver T. D. Human cranial anatomy and the differential preservation of population history and climate signatures. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 288, 1225–1233 (2006). - PubMed
-
- von Cramon-Taubadel N. Evolutionary insights into global patterns of human cranial diversity: population history, climatic and dietary effects. J. Anthropol. Sci. 92, 43–77 (2014). - PubMed
-
- Ritz-Timme S. et al. A new atlas for the evaluation of facial features: advantages, limits, and applicability. Int. J. Legal. Med. 125, 301–306 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

