Ribosome-based quality control of mRNA and nascent peptides
- PMID: 27193249
- PMCID: PMC5116004
- DOI: 10.1002/wrna.1366
Ribosome-based quality control of mRNA and nascent peptides
Abstract
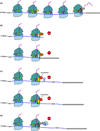
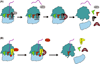
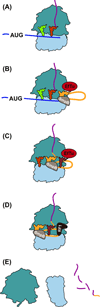
Quality control processes are widespread and play essential roles in detecting defective molecules and removing them in order to maintain organismal fitness. Aberrant messenger RNA (mRNA) molecules, unless properly managed, pose a significant hurdle to cellular proteostasis. Often mRNAs harbor premature stop codons, possess structures that present a block to the translational machinery, or lack stop codons entirely. In eukaryotes, the three cytoplasmic mRNA-surveillance processes, nonsense-mediated decay (NMD), no-go decay (NGD), and nonstop decay (NSD), evolved to cope with these aberrant mRNAs, respectively. Nonstop mRNAs and mRNAs that inhibit translation elongation are especially problematic as they sequester valuable ribosomes from the translating ribosome pool. As a result, in addition to RNA degradation, NSD and NGD are intimately coupled to ribosome rescue in all domains of life. Furthermore, protein products produced from all three classes of defective mRNAs are more likely to malfunction. It is not surprising then that these truncated nascent protein products are subject to degradation. Over the past few years, many studies have begun to document a central role for the ribosome in initiating the RNA and protein quality control processes. The ribosome appears to be responsible for recognizing the target mRNAs as well as for recruiting the factors required to carry out the processes of ribosome rescue and nascent protein decay. WIREs RNA 2017, 8:e1366. doi: 10.1002/wrna.1366 For further resources related to this article, please visit the WIREs website.
© 2016 Wiley Periodicals, Inc.
Figures


References
-
- Wolff S, Weissman JS, Dillin A. Differential Scales of Protein Quality Control. Cell. 2014;157:52–64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

