A population-based temporal logic gate for timing and recording chemical events
- PMID: 27193783
- PMCID: PMC5289221
- DOI: 10.15252/msb.20156663
A population-based temporal logic gate for timing and recording chemical events
Abstract
Engineered bacterial sensors have potential applications in human health monitoring, environmental chemical detection, and materials biosynthesis. While such bacterial devices have long been engineered to differentiate between combinations of inputs, their potential to process signal timing and duration has been overlooked. In this work, we present a two-input temporal logic gate that can sense and record the order of the inputs, the timing between inputs, and the duration of input pulses. Our temporal logic gate design relies on unidirectional DNA recombination mediated by bacteriophage integrases to detect and encode sequences of input events. For an E. coli strain engineered to contain our temporal logic gate, we compare predictions of Markov model simulations with laboratory measurements of final population distributions for both step and pulse inputs. Although single cells were engineered to have digital outputs, stochastic noise created heterogeneous single-cell responses that translated into analog population responses. Furthermore, when single-cell genetic states were aggregated into population-level distributions, these distributions contained unique information not encoded in individual cells. Thus, final differentiated sub-populations could be used to deduce order, timing, and duration of transient chemical events.
Keywords: DNA memory; event detectors; integrases; population analysis; stochastic biomolecular models.
© 2016 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
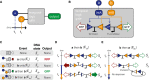
A temporal logic gate distinguishes between two chemical inputs (a, b) with different start times.
Implementation of the temporal logic gate using a set of two integrases with overlapping attachment sites. Chemical inputs a and b activate production of integrases intA and intB, which act upon a chromosomal DNA cassette.
Table with all possible inputs and outcomes to the event detector.
Sequence of DNA flipping following inputs with inducer a before inducer b (event E ab).
Sequence of DNA flipping following inducer inputs with b first (event E ba). In any events in which b precedes a, the unidirectionality of the intB attachment sites results in excision.
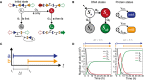
The four possible DNA states, illustrated with DNA state diagrams. All DNA begins in the initial state S o, and there are no reverse processes. The propensity functions α1, α2, and α3 are dependent on the concentration of the two integrases and correspond to the events b first (E b), a only (E a), and a then b (E ab), respectively.
Representation of the same model as a Markov chain. Integrases are represented simply as protein states with production (γA; γB) and degradation (δA; δB) rates.
Graphical representation of inducer step functions. ∆t is defined as difference between the start time of the first inducer and start time of the second.
Simulation results for inducer separation times of 0 and 5 h. There are four possible DNA states, but all cells end up in either the S b or S ab final states. Individual trajectories are simulated for 5,000 cells and the number of cells in each DNA state is summed for each time point (Appendix Fig S2).

The population fraction (N/5,000 cells) that switches into state S ab following an E ab event is dependent on the inducer separation time, ∆t. The gray to dark green color gradient represents increasing ∆t values. Square markers indicate final population fractions for specific values of ∆t.
In the case of the inverse E ba event, the fraction of cells in state S ab decreases monotonically with increasing ∆t. Circular markers indicate final population fractions for specific values of ∆t.
Final S ab cell fractions from (A, B) are plotted as a function of ∆t. Blue line with square markers shows end point population fractions from an E ab event. Yellow line with circular markers shows final end point population fractions from an E ba event. The gradient inside the markers corresponds to increasing ∆t value. The dotted gray line corresponds to the ∆t 90, the value of ∆t at which ≥ 90% of the cells are in state S ab. All simulations were done with a population of N = 5,000 cells.
Chart showing differences in information that can be recorded at the single‐cell versus the population level. In particular, E ba does not have a unique single‐cell genetic state, but has a clear distinct population‐level phenotype.

Populations of cells exposed to an E ab event sequence. Cell switching to state S ab (indicated by GFP fluorescence) begins when inducer b (aTc) is added. Maximum normalized GFP fluorescence increases as a function of the inducer separation time ∆t. Gray to dark green gradient represents increasing ∆t values. Square markers are final end point measurements. Error bars represent standard error of the mean.
Cells exposed to the inverse E ba sequence of events. GFP fluorescence decreases monotonically with increasing inducer separation time between b and a. Circular markers are final end point measurements.
Final population distributions from (A, B) at 30 h are plotted as a function of ∆t. Cells were gated by GFP fluorescence to identify percentage of S ab cells. Dotted line marks ∆t 90 detection limit.
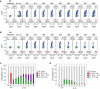
E ab cell populations plotted by their RFP and GFP expression with increasing ∆t. Leaky expression of PBAD‐intA can be estimated by looking at Q3 of the No inducers,b only populations (˜0.5–2%). Leaky expression of Ptet‐intB can be estimated with Q1 + Q2 fractions of the a only population (˜2–3%).
E ba populations with increasing ∆t.
Population fractions by quadrant for a then b, E ab.
Population fractions by quadrant for b then a, E ba. Individual flow cytometry histograms can be found in Appendix Figs S8–S10.

As DNA flipping rates of intA () are decreased relative to , the population of S ab cells at ∆t = 0 h has a downward shift. Simulations are done with N = 3,000 trajectories/marker.
Increasing the leaky expression of intB () changes the maximum threshold of cells that correctly identify S ab even at high ∆t. Leakiness is defined as a percentage of the induced integrase production rate ().
The model was revised to more closely match the experimental data by constraining parameters for leaky expression and varying integrase flipping (N = 5,000). Mean squared error was calculated between the experimental data and the initial and revised models to find an optimized pair of values (Appendix Fig S13). The revised parameters are k flipA = 0.2 h−1, k flipB = 0.3 h−1, k leakA = 1% of k prodA(μm3 · h)−1, and k leakB = 2% of k prodB(μm3 · h)−1.

Inducer a can be used as a reference signal against which to measure the time and duration of the inducer b pulse.
The population eventually divides into one of two sub‐populations: those that see inducer a first and those that see inducer b first. Only if a cell has entered the a first pathway does it have the possibility to express RFP or GFP. Furthermore, S a can be thought of as a necessary precursor to S ab.
A matrix illustrating a subset of the ∆t and PW b values to be tested.
Simulation results show that for any given ∆t, the number of cells in S b = (total number of cells − (S a + S ab))
The fraction of the population in the S a state is totally independent of ∆t and depends only on the pulse duration of inducer b.
Once PW b is known, then the fraction of the population in S ab state can be used to find the time at which the pulse of inducer b began. N = 3,000 cell trajectories for each value of ∆t, PW b.
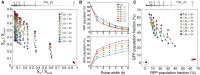
Simulation results from testing an 11 × 11 matrix of parameters with ∆t and PW b varying from 0 to 6 h in increments of 0.5 h. Each point represents a population of 3,000 cells. Increasing PW b goes from right to left, and increasing ∆t goes from bottom to top.
Experimental results showing RFP and GFP population fractions as a function of increasing ∆t and PW b. Experimental results were obtained by exposing temporal logic gate E. coli populations to varying PW b and ∆t values (0–6 h, 0.01%/vol L‐ara, 200 ng/ml aTc, measurements taken at 48 h).
A scatterplot of each population using their RFP and GFP fractions as coordinates (˜106 cells per population). The non‐induced control samples are indicated with a dotted circle on the bottom left, and the samples with PW b = 0 h are on the bottom right. Samples with the same PW b are connected with a solid line, and line darkness represents increasing PW b duration. Samples with the same ∆t are shown with the same colored shape marker and increasing ∆t goes from bottom to top.

Control population not exposed to any inducers. There is minimal leaky expression (1–2%) into Q3 after 36 h of growth.
Populations for PW b and ∆t values of 0, 3, and 6 h. For PW b = 0 h populations, ˜60% of the cells switch to S a (Q3), with ˜3% intB leaky expression going into Q1 and Q2. As PW b increases, the S a fraction drops from 60% (PW b = 0 h) to 10–20% (PW b = 3 h) to less than 10% (PW b = 6 h). As ∆t increases, the percentage of cells in S ab (Q1) increases from 20–40% (∆t = 0 h) to 40–50% (∆t = 3 h) to 50–60% (∆t = 6 h). S a populations (Q3) also drift downwards with increasing ∆t, rather than staying constant as predicted in simulation. Lower ∆t results in higher S b populations, which, combined with S o cells, make up Q4. Critically, the percentage of the population expressing both RFP and GFP simultaneously (Q2) is always < 3%. This ensures that RFP is a reliable determinant of S a state cells, and subsequently, of PW b.
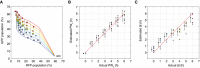
A mesh generated from fitted curves for PW b as a function of RFP population percentage(R) and ∆t as a function of pulse width and GFP population percentage(G). Experimental data are overlaid.
Comparison of actual versus estimated PW b values generated by fitted function PW b(R). For each actual PW b value, the average of the estimated PW b values with ± 1 standard deviation (slightly offset on the x‐axis for better comparison).
Comparison of actual versus estimated ∆t generated by the fitted function ∆t(G; PW b). For each actual ∆t value, the average of the estimated ∆t with ± 1 standard deviation (slightly offset on the x‐axis for better comparison).
Similar articles
-
Amplifying genetic logic gates.Science. 2013 May 3;340(6132):599-603. doi: 10.1126/science.1232758. Epub 2013 Mar 28. Science. 2013. PMID: 23539178
-
Processing two environmental chemical signals with a synthetic genetic IMPLY gate, a 2-input-2-output integrated logic circuit, and a process pipeline to optimize its systems chemistry in Escherichia coli.Biotechnol Bioeng. 2020 May;117(5):1502-1512. doi: 10.1002/bit.27286. Epub 2020 Feb 7. Biotechnol Bioeng. 2020. PMID: 31981217
-
Design, Fabrication, and Device Chemistry of a 3-Input-3-Output Synthetic Genetic Combinatorial Logic Circuit with a 3-Input AND Gate in a Single Bacterial Cell.Bioconjug Chem. 2019 Dec 18;30(12):3013-3020. doi: 10.1021/acs.bioconjchem.9b00517. Epub 2019 Oct 24. Bioconjug Chem. 2019. PMID: 31596072
-
Phage integrases: biology and applications.J Mol Biol. 2004 Jan 16;335(3):667-78. doi: 10.1016/j.jmb.2003.09.082. J Mol Biol. 2004. PMID: 14687564 Review.
-
Making serine integrases work for us.Curr Opin Microbiol. 2017 Aug;38:130-136. doi: 10.1016/j.mib.2017.04.006. Epub 2017 Jun 6. Curr Opin Microbiol. 2017. PMID: 28599144 Review.
Cited by
-
Untethered Soft Microrobots with Adaptive Logic Gates.Adv Sci (Weinh). 2023 May;10(13):e2206662. doi: 10.1002/advs.202206662. Epub 2023 Feb 21. Adv Sci (Weinh). 2023. PMID: 36809583 Free PMC article.
-
Scaling up genetic circuit design for cellular computing: advances and prospects.Nat Comput. 2018;17(4):833-853. doi: 10.1007/s11047-018-9715-9. Epub 2018 Oct 5. Nat Comput. 2018. PMID: 30524216 Free PMC article.
-
An Exo III-powered closed-loop DNA circuit architecture for biosensing/imaging.Mikrochim Acta. 2024 Jun 14;191(7):395. doi: 10.1007/s00604-024-06476-0. Mikrochim Acta. 2024. PMID: 38877347
-
Molecular recording: transcriptional data collection into the genome.Curr Opin Biotechnol. 2023 Feb;79:102855. doi: 10.1016/j.copbio.2022.102855. Epub 2022 Dec 5. Curr Opin Biotechnol. 2023. PMID: 36481341 Free PMC article. Review.
-
Ultrasound-controllable engineered bacteria for cancer immunotherapy.Nat Commun. 2022 Mar 24;13(1):1585. doi: 10.1038/s41467-022-29065-2. Nat Commun. 2022. PMID: 35332124 Free PMC article.
References
-
- Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D (2013) Amplifying genetic logic gates. Science 340: 599–603 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

