Species-level core oral bacteriome identified by 16S rRNA pyrosequencing in a healthy young Arab population
- PMID: 27193835
- PMCID: PMC4871898
- DOI: 10.3402/jom.v8.31444
Species-level core oral bacteriome identified by 16S rRNA pyrosequencing in a healthy young Arab population
Abstract
Background: Reports on the composition of oral bacteriome in Arabs are lacking. In addition, the majority of previous studies on other ethnic groups have been limited by low-resolution taxonomic assignment of next-generation sequencing reads. Furthermore, there has been a conflict about the existence of a 'core' bacteriome.
Objective: The objective of this study was to characterize the healthy core oral bacteriome in a young Arab population at the species level.
Methods: Oral rinse DNA samples obtained from 12 stringently selected healthy young subjects of Arab origin were pyrosequenced (454's FLX chemistry) for the bacterial 16S V1-V3 hypervariable region at an average depth of 11,500 reads. High-quality, non-chimeric reads ≥380 bp were classified to the species level using the recently described, prioritized, multistage assignment algorithm. A core bacteriome was defined as taxa present in at least 11 samples. The Chao2, abundance-based coverage estimator (ACE), and Shannon indices were computed to assess species richness and diversity.
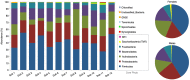
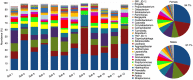
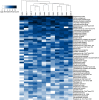
Results: Overall, 557 species-level taxa (211±42 per subject) were identified, representing 122 genera and 13 phyla. The core bacteriome comprised 55 species-level taxa belonging to 30 genera and 7 phyla, namely Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, Fusobacteria, Saccharibacteria, and SR1. The core species constituted between 67 and 87% of the individual bacteriomes. However, the abundances differed by up to three orders of magnitude among the study subjects. On average, Streptococcus mitis, Rothia mucilaginosa, Haemophilus parainfluenzae, Neisseria flavescence/subflava group, Prevotella melaninogenica, and Veillonella parvula group were the most abundant. Streptococcus sp. C300, a taxon never reported in the oral cavity, was identified as a core species. Species richness was estimated at 586 (Chao2) and 614 (ACE) species, whereas diversity (Shannon index) averaged at 3.99.
Conclusions: A species-level core oral bacteriome representing the majority of reads was identified, which can serve as a reference for comparison with oral bacteriomes of other populations as well as those associated with disease.
Keywords: core; high-throughput nucleotide sequencing; human; micgrobiome; mouth; pyrosequencing.
Figures
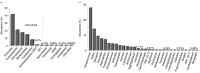
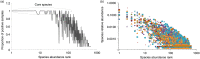

References
-
- Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149:279–94. - PubMed
-
- Siqueira JF, Jr, Fouad AF, Rocas IN. Pyrosequencing as a tool for better understanding of human microbiomes. J Oral Microbiol. 2012;4 10743, doi: http://dx.doi.org/10.3402/jom.v4i0.10743. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
