Effect of salivary agglutination on oral streptococcal clearance by human polymorphonuclear neutrophil granulocytes
- PMID: 27194631
- PMCID: PMC5116291
- DOI: 10.1111/omi.12164
Effect of salivary agglutination on oral streptococcal clearance by human polymorphonuclear neutrophil granulocytes
Abstract
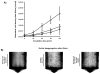
Salivary agglutination is an important host defense mechanism to aggregate oral commensal bacteria as well as invading pathogens. Saliva flow and subsequent swallowing more easily clear aggregated bacteria compared with single cells. Phagocytic clearance of bacteria through polymorphonuclear neutrophil granulocytes also seems to increase to a certain extent with the size of bacterial aggregates. To determine a connection between salivary agglutination and the host innate immune response by phagocytosis, an in vitro agglutination assay was developed reproducing the average size of salivary bacterial aggregates. Using the oral commensal Streptococcus gordonii as a model organism, the effect of salivary agglutination on phagocytic clearance through polymorphonuclear neutrophil granulocytes was investigated. Here we describe how salivary aggregates of S. gordonii are readily cleared through phagocytosis, whereas single bacterial cells showed a significant delay in being phagocytosed and killed. Furthermore, before phagocytosis the polymorphonuclear neutrophil granulocytes were able to induce a specific de-aggregation, which was dependent on serine protease activity. The data presented suggest that salivary agglutination of bacterial cells leads to an ideal size for recognition by polymorphonuclear neutrophil granulocytes. As a first line of defense, these phagocytic cells are able to recognize the aggregates and de-aggregate them via serine proteases to a more manageable size for efficient phagocytosis and subsequent killing in the phagolysosome. This observed mechanism not only prevents the rapid spreading of oral bacterial cells while entering the bloodstream but would also avoid degranulation of involved polymorphonuclear neutrophil granulocytes, so preventing collateral damage to nearby tissue.
Keywords: Streptococcus gordonii; innate immunity; polymorphonuclear neutrophil granulocytes; salivary agglutination.
© 2016 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

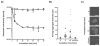

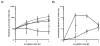
Similar articles
-
Phagocytic and killing activity of human blood, gingival crevicular, and salivary polymorphonuclear leukocytes for oral streptococci.J Dent Res. 1982 May;61(5):636-9. doi: 10.1177/00220345820610050301. J Dent Res. 1982. PMID: 7045181
-
Contribution of phosphoglucosamine mutase to the resistance of Streptococcus gordonii DL1 to polymorphonuclear leukocyte killing.FEMS Microbiol Lett. 2009 Aug;297(2):196-202. doi: 10.1111/j.1574-6968.2009.01673.x. Epub 2009 Jun 3. FEMS Microbiol Lett. 2009. PMID: 19552711
-
Effect of co-aggregation on the pathogenicity of oral bacteria.J Med Microbiol. 1993 Sep;39(3):183-90. doi: 10.1099/00222615-39-3-183. J Med Microbiol. 1993. PMID: 8366516
-
Neutrophil granulocytes as host cells and transport vehicles for intracellular pathogens: apoptosis as infection-promoting factor.Immunobiology. 2008;213(3-4):183-91. doi: 10.1016/j.imbio.2007.11.010. Epub 2008 Feb 8. Immunobiology. 2008. PMID: 18406366 Review.
-
Neutrophils and aquatic pathogens.Parasite Immunol. 2022 Jun;44(6):e12915. doi: 10.1111/pim.12915. Epub 2022 Mar 22. Parasite Immunol. 2022. PMID: 35290688 Free PMC article. Review.
Cited by
-
Pathophysiological Responses of Oral Keratinocytes After Exposure to Flavored E-Cigarette Liquids.Dent J (Basel). 2025 Jan 29;13(2):60. doi: 10.3390/dj13020060. Dent J (Basel). 2025. PMID: 39996934 Free PMC article.
-
Corynebacterial membrane vesicles disrupt cariogenic interkingdom assemblages.Appl Environ Microbiol. 2024 Nov 20;90(11):e0088524. doi: 10.1128/aem.00885-24. Epub 2024 Oct 31. Appl Environ Microbiol. 2024. PMID: 39480093 Free PMC article.
-
Impact of Polymicrobial Infection on Fitness of Streptococcus gordonii In Vivo.mBio. 2023 Jun 27;14(3):e0065823. doi: 10.1128/mbio.00658-23. Epub 2023 Apr 12. mBio. 2023. PMID: 37042761 Free PMC article.
-
Effects of equine SALSA on neutrophil phagocytosis and macrophage cytokine production.PLoS One. 2022 Mar 14;17(3):e0264911. doi: 10.1371/journal.pone.0264911. eCollection 2022. PLoS One. 2022. PMID: 35286327 Free PMC article.
-
Glycan recognition at the saliva - oral microbiome interface.Cell Immunol. 2018 Nov;333:19-33. doi: 10.1016/j.cellimm.2018.08.008. Epub 2018 Aug 18. Cell Immunol. 2018. PMID: 30274839 Free PMC article. Review.
References
-
- Amerongen AV, Veerman EC. Saliva-the defender of the oral cavity. Oral Dis. 2002;8:12–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

