A New Genomics-Driven Taxonomy of Bacteria and Archaea: Are We There Yet?
- PMID: 27194687
- PMCID: PMC4963521
- DOI: 10.1128/JCM.00200-16
A New Genomics-Driven Taxonomy of Bacteria and Archaea: Are We There Yet?
Abstract
Taxonomy is often criticized for being too conservative and too slow and having limited relevance because it has not taken into consideration the latest methods and findings. Yet the cumulative work product of its practitioners underpins contemporary microbiology and serves as a principal means of shaping and referencing knowledge. Using methods drawn from the field of exploratory data analysis, this minireview examines the current state of the field as it transitions from a taxonomy based on 16S rRNA gene sequences to one based on whole-genome sequences and tests the validity of some commonly held beliefs.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
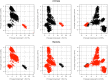
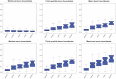

References
-
- Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kampfer P, Maiden MC, Nesme X, Rossello-Mora R, Swings J, Truper HG, Vauterin L, Ward AC, Whitman WB. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047. doi:10.1099/00207713-52-3-1043. - DOI - PubMed
-
- Yarza P, Sproer C, Swiderski J, Mrotzek N, Spring S, Tindall BJ, Gronow S, Pukall R, Klenk HP, Lang E, Verbarg S, Crouch A, Lilburn T, Beck B, Unosson C, Cardew S, Moore ER, Gomila M, Nakagawa Y, Janssens D, De Vos P, Peiren J, Suttels T, Clermont D, Bizet C, Sakamoto M, Iida T, Kudo T, Kosako Y, Oshida Y, Ohkuma M, Arahal DR, Spieck E, Pommerening Roeser A, Figge M, Park D, Buchanan P, Cifuentes A, Munoz R, Euzeby JP, Schleifer KH, Ludwig W, Amann R, Glockner FO, Rossello-Mora R. 2013. Sequencing orphan species initiative (SOS): filling the gaps in the 16S rRNA gene sequence database for all species with validly published names. Syst Appl Microbiol 36:69–73. doi:10.1016/j.syapm.2012.12.006. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

