Both cis and trans Activities of Foot-and-Mouth Disease Virus 3D Polymerase Are Essential for Viral RNA Replication
- PMID: 27194768
- PMCID: PMC4944275
- DOI: 10.1128/JVI.00469-16
Both cis and trans Activities of Foot-and-Mouth Disease Virus 3D Polymerase Are Essential for Viral RNA Replication
Abstract
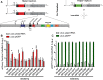
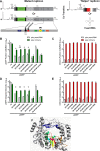
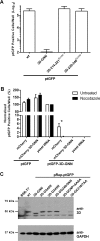
The Picornaviridae is a large family of positive-sense RNA viruses that contains numerous human and animal pathogens, including foot-and-mouth disease virus (FMDV). The picornavirus replication complex comprises a coordinated network of protein-protein and protein-RNA interactions involving multiple viral and host-cellular factors. Many of the proteins within the complex possess multiple roles in viral RNA replication, some of which can be provided in trans (i.e., via expression from a separate RNA molecule), while others are required in cis (i.e., expressed from the template RNA molecule). In vitro studies have suggested that multiple copies of the RNA-dependent RNA polymerase (RdRp) 3D are involved in the viral replication complex. However, it is not clear whether all these molecules are catalytically active or what other function(s) they provide. In this study, we aimed to distinguish between catalytically active 3D molecules and those that build a replication complex. We report a novel nonenzymatic cis-acting function of 3D that is essential for viral-genome replication. Using an FMDV replicon in complementation experiments, our data demonstrate that this cis-acting role of 3D is distinct from the catalytic activity, which is predominantly trans acting. Immunofluorescence studies suggest that both cis- and trans-acting 3D molecules localize to the same cellular compartment. However, our genetic and structural data suggest that 3D interacts in cis with RNA stem-loops that are essential for viral RNA replication. This study identifies a previously undescribed aspect of picornavirus replication complex structure-function and an important methodology for probing such interactions further.
Importance: Foot-and-mouth disease virus (FMDV) is an important animal pathogen responsible for foot-and-mouth disease. The disease is endemic in many parts of the world with outbreaks within livestock resulting in major economic losses. Propagation of the viral genome occurs within replication complexes, and understanding this process can facilitate the development of novel therapeutic strategies. Many of the nonstructural proteins involved in replication possess multiple functions in the viral life cycle, some of which can be supplied to the replication complex from a separate genome (i.e., in trans) while others must originate from the template (i.e., in cis). Here, we present an analysis of cis and trans activities of the RNA-dependent RNA polymerase 3D. We demonstrate a novel cis-acting role of 3D in replication. Our data suggest that this role is distinct from its enzymatic functions and requires interaction with the viral genome. Our data further the understanding of genome replication of this important pathogen.
Copyright © 2016 Herod et al.
Figures



Similar articles
-
Insights into Polyprotein Processing and RNA-Protein Interactions in Foot-and-Mouth Disease Virus Genome Replication.J Virol. 2023 May 31;97(5):e0017123. doi: 10.1128/jvi.00171-23. Epub 2023 May 8. J Virol. 2023. PMID: 37154761 Free PMC article.
-
Heat shock protein A1 inhibits the replication of foot-and-mouth disease virus by degrading viral RNA polymerase 3D through chaperone-mediated autophagy.J Virol. 2025 May 20;99(5):e0016825. doi: 10.1128/jvi.00168-25. Epub 2025 Mar 31. J Virol. 2025. PMID: 40162788 Free PMC article.
-
Attenuation of Foot-and-Mouth Disease Virus by Engineered Viral Polymerase Fidelity.J Virol. 2017 Jul 12;91(15):e00081-17. doi: 10.1128/JVI.00081-17. Print 2017 Aug 1. J Virol. 2017. PMID: 28515297 Free PMC article.
-
The multiple roles of viral 3Dpol protein in picornavirus infections.Virulence. 2024 Dec;15(1):2333562. doi: 10.1080/21505594.2024.2333562. Epub 2024 Apr 15. Virulence. 2024. PMID: 38622757 Free PMC article. Review.
-
Structural insights into replication initiation and elongation processes by the FMDV RNA-dependent RNA polymerase.Curr Opin Struct Biol. 2009 Dec;19(6):752-8. doi: 10.1016/j.sbi.2009.10.016. Epub 2009 Nov 13. Curr Opin Struct Biol. 2009. PMID: 19914060 Review.
Cited by
-
Insights into Polyprotein Processing and RNA-Protein Interactions in Foot-and-Mouth Disease Virus Genome Replication.J Virol. 2023 May 31;97(5):e0017123. doi: 10.1128/jvi.00171-23. Epub 2023 May 8. J Virol. 2023. PMID: 37154761 Free PMC article.
-
The DEAD-Box RNA Helicase DDX1 Interacts with the Viral Protein 3D and Inhibits Foot-and-Mouth Disease Virus Replication.Virol Sin. 2019 Dec;34(6):610-617. doi: 10.1007/s12250-019-00148-7. Epub 2019 Jul 29. Virol Sin. 2019. PMID: 31359346 Free PMC article.
-
A Novel Plasmid DNA-Based Foot and Mouth Disease Virus Minigenome for Intracytoplasmic mRNA Production.Viruses. 2021 Jun 1;13(6):1047. doi: 10.3390/v13061047. Viruses. 2021. PMID: 34205958 Free PMC article.
-
Functional advantages of triplication of the 3B coding region of the FMDV genome.FASEB J. 2021 Feb;35(2):e21215. doi: 10.1096/fj.202001473RR. Epub 2020 Nov 23. FASEB J. 2021. PMID: 33230899 Free PMC article.
-
Genetic economy in picornaviruses: Foot-and-mouth disease virus replication exploits alternative precursor cleavage pathways.PLoS Pathog. 2017 Oct 2;13(10):e1006666. doi: 10.1371/journal.ppat.1006666. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 28968463 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- BB/F01614X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- C20035/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0901002/MRC_/Medical Research Council/United Kingdom
- BB/K003801/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E010709/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

