The calmodulin-binding transcription activator CAMTA1 is required for long-term memory formation in mice
- PMID: 27194798
- PMCID: PMC4880143
- DOI: 10.1101/lm.041111.115
The calmodulin-binding transcription activator CAMTA1 is required for long-term memory formation in mice
Abstract
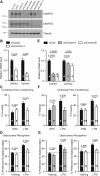
The formation of long-term memory requires signaling from the synapse to the nucleus to mediate neuronal activity-dependent gene transcription. Synapse-to-nucleus communication is initiated by influx of calcium ions through synaptic NMDA receptors and/or L-type voltage-gated calcium channels and involves the activation of transcription factors by calcium/calmodulin signaling in the nucleus. Recent studies have drawn attention to a new family of transcriptional regulators, the so-called calmodulin-binding transcription activator (CAMTA) proteins. CAMTAs are expressed at particularly high levels in the mouse and human brain, and we reasoned that, as calmodulin-binding transcription factors, CAMTAs may regulate the formation of long-term memory by coupling synaptic activity and calcium/calmodulin signaling to memory-related transcriptional responses. This hypothesis is supported by genetic studies that reported a correlation between Camta gene polymorphisms or mutations and cognitive capability in humans. Here, we show that acute knockdown of CAMTA1, but not CAMTA2, in the hippocampus of adult mice results in impaired performance in two memory tests, contextual fear conditioning and object-place recognition test. Short-term memory and neuronal morphology were not affected by CAMTA knockdown. Gene expression profiling in the hippocampus of control and CAMTA knockdown mice revealed a number of putative CAMTA1 target genes related to synaptic transmission and neuronal excitability. Patch clamp recordings in organotypic hippocampal slice cultures provided further evidence for CAMTA1-dependent changes in electrophysiological properties. In summary, our study provides experimental evidence that confirms previous human genetic studies and establishes CAMTA1 as a regulator of long-term memory formation.
© 2016 Bas-Orth et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Bading H. 2013. Nuclear calcium signalling in the regulation of brain function. Nat Rev Neurosci 14: 593–608. - PubMed
-
- Bading H, Greenberg ME. 1991. Stimulation of protein tyrosine phosphorylation by NMDA receptor activation. Science 253: 912–914. - PubMed
-
- Bading H, Ginty DD, Greenberg ME. 1993. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science 260: 181–186. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
