Construction and Annotation of a High Density SNP Linkage Map of the Atlantic Salmon (Salmo salar) Genome
- PMID: 27194803
- PMCID: PMC4938670
- DOI: 10.1534/g3.116.029009
Construction and Annotation of a High Density SNP Linkage Map of the Atlantic Salmon (Salmo salar) Genome
Abstract
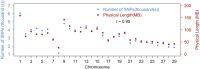
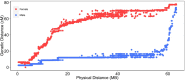
High density linkage maps are useful tools for fine-scale mapping of quantitative trait loci, and characterization of the recombination landscape of a species' genome. Genomic resources for Atlantic salmon (Salmo salar) include a well-assembled reference genome, and high density single nucleotide polymorphism (SNP) arrays. Our aim was to create a high density linkage map, and to align it with the reference genome assembly. Over 96,000 SNPs were mapped and ordered on the 29 salmon linkage groups using a pedigreed population comprising 622 fish from 60 nuclear families, all genotyped with the 'ssalar01' high density SNP array. The number of SNPs per group showed a high positive correlation with physical chromosome length (r = 0.95). While the order of markers on the genetic and physical maps was generally consistent, areas of discrepancy were identified. Approximately 6.5% of the previously unmapped reference genome sequence was assigned to chromosomes using the linkage map. Male recombination rate was lower than females across the vast majority of the genome, but with a notable peak in subtelomeric regions. Finally, using RNA-Seq data to annotate the reference genome, the mapped SNPs were categorized according to their predicted function, including annotation of ∼2500 putative nonsynonymous variants. The highest density SNP linkage map for any salmonid species has been created, annotated, and integrated with the Atlantic salmon reference genome assembly. This map highlights the marked heterochiasmy of salmon, and provides a useful resource for salmonid genetics and genomics research.
Keywords: RNA-Seq; SNP array; Salmo salar; linkage map; recombination.
Copyright © 2016 Tsai et al.
Figures

Similar articles
-
Linkage maps of the Atlantic salmon (Salmo salar) genome derived from RAD sequencing.BMC Genomics. 2014 Feb 27;15:166. doi: 10.1186/1471-2164-15-166. BMC Genomics. 2014. PMID: 24571138 Free PMC article.
-
Development and validation of a high density SNP genotyping array for Atlantic salmon (Salmo salar).BMC Genomics. 2014 Feb 6;15:90. doi: 10.1186/1471-2164-15-90. BMC Genomics. 2014. PMID: 24524230 Free PMC article.
-
Characterisation of QTL-linked and genome-wide restriction site-associated DNA (RAD) markers in farmed Atlantic salmon.BMC Genomics. 2012 Jun 15;13:244. doi: 10.1186/1471-2164-13-244. BMC Genomics. 2012. PMID: 22702806 Free PMC article.
-
A linkage map of the Atlantic salmon (Salmo salar) based on EST-derived SNP markers.BMC Genomics. 2008 May 15;9:223. doi: 10.1186/1471-2164-9-223. BMC Genomics. 2008. PMID: 18482444 Free PMC article.
-
Atlantic salmon (Salmo salar L.) genetics in the 21st century: taking leaps forward in aquaculture and biological understanding.Anim Genet. 2019 Feb;50(1):3-14. doi: 10.1111/age.12748. Epub 2018 Nov 14. Anim Genet. 2019. PMID: 30426521 Free PMC article. Review.
Cited by
-
New Paradigms to Help Solve the Global Aquaculture Disease Crisis.PLoS Pathog. 2017 Feb 2;13(2):e1006160. doi: 10.1371/journal.ppat.1006160. eCollection 2017 Feb. PLoS Pathog. 2017. PMID: 28152043 Free PMC article. No abstract available.
-
A Dense Brown Trout (Salmo trutta) Linkage Map Reveals Recent Chromosomal Rearrangements in the Salmo Genus and the Impact of Selection on Linked Neutral Diversity.G3 (Bethesda). 2017 Apr 3;7(4):1365-1376. doi: 10.1534/g3.116.038497. G3 (Bethesda). 2017. PMID: 28235829 Free PMC article.
-
Applications of genotyping by sequencing in aquaculture breeding and genetics.Rev Aquac. 2018 Aug;10(3):670-682. doi: 10.1111/raq.12193. Epub 2017 Feb 4. Rev Aquac. 2018. PMID: 30220910 Free PMC article. Review.
-
Highly dense linkage maps from 31 full-sibling families of turbot (Scophthalmus maximus) provide insights into recombination patterns and chromosome rearrangements throughout a newly refined genome assembly.DNA Res. 2018 Aug 1;25(4):439-450. doi: 10.1093/dnares/dsy015. DNA Res. 2018. PMID: 29897548 Free PMC article.
-
High-Density Linkage Map and QTLs for Growth in Snapper (Chrysophrys auratus).G3 (Bethesda). 2019 Apr 9;9(4):1027-1035. doi: 10.1534/g3.118.200905. G3 (Bethesda). 2019. PMID: 30804023 Free PMC article.
References
-
- Allendorf, F. W., and G. H. Thorgaard, 1984 Evolutionary genetics of fishes, pp. 55–93 in Tetraploidy and the Evolution of Salmonid Fishes, edited by B. J. Turner, Plenum Press, New York.
-
- Allendorf F. W., Bassham S., Cresko W. A., Limborg M. T., Seeb L. W., et al. , 2015. Effects of crossovers between homeologs on inheritance and population genomics in polyploid-derived salmonid fishes. J. Hered. 106: 217–227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/F001959/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F002750/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H022007/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/K001744/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
