Smaug variants in neural and non-neuronal cells
- PMID: 27195061
- PMCID: PMC4857778
- DOI: 10.1080/19420889.2016.1139252
Smaug variants in neural and non-neuronal cells
Abstract
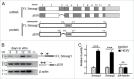
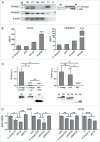
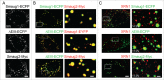
Mammalian Smaug1/Samd4a is an mRNA regulator involved in synapse plasticity and additional non-neuronal functions. Here we analyzed the expression of Smaug1/Samd4a variants and Smaug2/Samd4b in primary hippocampal neurons and non-neuronal cell lines. We found that multiple Smaug proteins are present in several mammalian cell lines, including a canonical full length Smaug1, a Smaug1 variant that lacks the third exon, termed ΔEIII, and Smaug2, the product of a highly homologous gene. These three major isoforms are expressed differentially along neuron development and form cytosolic bodies when transfected in cell lines. By using luciferase reporters, we found that the ΔEIII isoform, which lacks 10 amino acids in the sterile α motif involved in RNA binding, shows a RNA-binding capacity and repressor activity comparable to that of the full length Smaug1. These observations are an important groundwork for molecular studies of the Smaug post-transcriptional pathway, which is relevant to neuron development, mitochondrial function and muscle physiology in health and disease.
Keywords: mRNA silencing; muscular dystrophy; processing bodies; stress granules; synaptogenesis; translation.
Figures
References
-
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal 2011; 23:324-34; PMID:20813183; http://dx.doi.org/ 10.1016/j.cellsig.2010.08.011 - DOI - PMC - PubMed
-
- Baez MV, Luchelli L, Maschi D, Habif M, Pascual M, Thomas MG, Boccaccio GL. Smaug1 mRNA-silencing foci respond to NMDA and modulate synapse formation. J Cell Biol 2011; 195:1141-57; PMID:22201125; http://dx.doi.org/ 10.1083/jcb.201108159 - DOI - PMC - PubMed
-
- Luchelli L, Thomas MG, Boccaccio GL. Synaptic control of mRNA translation by reversible assembly of XRN1 bodies. J Cell Sci 2015; 128:1542-54; PMID:25736288; http://dx.doi.org/ 10.1242/jcs.163295 - DOI - PubMed
-
- Pascual ML, Luchelli L, Habif M, Boccaccio GL. Synaptic activity regulated mRNA-silencing foci for the fine tuning of local protein synthesis at the synapse. Commun Integr Biol 2012; 5:388-92; PMID:23060966; http://dx.doi.org/ 10.4161/cib.20257 - DOI - PMC - PubMed
-
- Chartier A, Klein P, Pierson S, Barbezier N, Gidaro T, Casas F, Carberry S, Dowling P, Maynadier L, Bellec M, et al.. Mitochondrial dysfunction reveals the role of mRNA poly(A) tail regulation in oculopharyngeal muscular dystrophy pathogenesis. PLoS Genet 2015; 11:e1005092; PMID:25816335; http://dx.doi.org/ 10.1371/journal.pgen.1005092 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
