Introducing BASE: the Biomes of Australian Soil Environments soil microbial diversity database
- PMID: 27195106
- PMCID: PMC4870752
- DOI: 10.1186/s13742-016-0126-5
Introducing BASE: the Biomes of Australian Soil Environments soil microbial diversity database
Erratum in
-
Erratum to: Introducing BASE: the Biomes of Australian Soil Environments soil microbial diversity database.Gigascience. 2017 May 1;6(5):1. doi: 10.1093/gigascience/gix021. Gigascience. 2017. PMID: 30137319 Free PMC article. No abstract available.
Abstract
Background: Microbial inhabitants of soils are important to ecosystem and planetary functions, yet there are large gaps in our knowledge of their diversity and ecology. The 'Biomes of Australian Soil Environments' (BASE) project has generated a database of microbial diversity with associated metadata across extensive environmental gradients at continental scale. As the characterisation of microbes rapidly expands, the BASE database provides an evolving platform for interrogating and integrating microbial diversity and function.
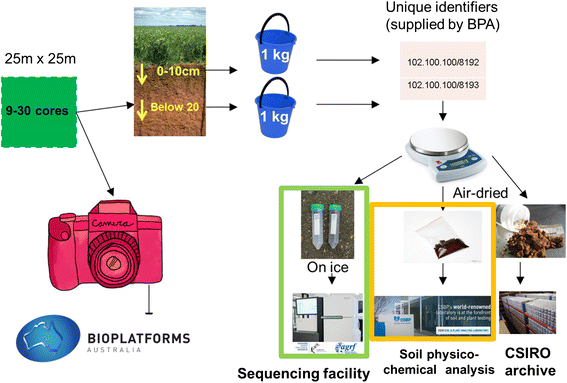
Findings: BASE currently provides amplicon sequences and associated contextual data for over 900 sites encompassing all Australian states and territories, a wide variety of bioregions, vegetation and land-use types. Amplicons target bacteria, archaea and general and fungal-specific eukaryotes. The growing database will soon include metagenomics data. Data are provided in both raw sequence (FASTQ) and analysed OTU table formats and are accessed via the project's data portal, which provides a user-friendly search tool to quickly identify samples of interest. Processed data can be visually interrogated and intersected with other Australian diversity and environmental data using tools developed by the 'Atlas of Living Australia'.
Conclusions: Developed within an open data framework, the BASE project is the first Australian soil microbial diversity database. The database will grow and link to other global efforts to explore microbial, plant, animal, and marine biodiversity. Its design and open access nature ensures that BASE will evolve as a valuable tool for documenting an often overlooked component of biodiversity and the many microbe-driven processes that are essential to sustain soil function and ecosystem services.
Keywords: Australia; Database; Metagenomics; Microbial diversity; Microbial ecology; Microbiology; Soil biology.
Figures


References
-
- Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat Rev Micro. 2012;10:497–506. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

