Challenges of Identifying Clinically Actionable Genetic Variants for Precision Medicine
- PMID: 27195526
- PMCID: PMC4955563
- DOI: 10.1155/2016/3617572
Challenges of Identifying Clinically Actionable Genetic Variants for Precision Medicine
Abstract
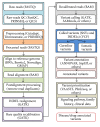
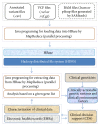
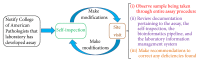
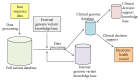
Advances in genomic medicine have the potential to change the way we treat human disease, but translating these advances into reality for improving healthcare outcomes depends essentially on our ability to discover disease- and/or drug-associated clinically actionable genetic mutations. Integration and manipulation of diverse genomic data and comprehensive electronic health records (EHRs) on a big data infrastructure can provide an efficient and effective way to identify clinically actionable genetic variants for personalized treatments and reduce healthcare costs. We review bioinformatics processing of next-generation sequencing (NGS) data, bioinformatics infrastructures for implementing precision medicine, and bioinformatics approaches for identifying clinically actionable genetic variants using high-throughput NGS data and EHRs.
Conflict of interest statement
The authors indicated no potential competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

