Ly6E/K Signaling to TGFβ Promotes Breast Cancer Progression, Immune Escape, and Drug Resistance
- PMID: 27197181
- PMCID: PMC4910623
- DOI: 10.1158/0008-5472.CAN-15-2654
Ly6E/K Signaling to TGFβ Promotes Breast Cancer Progression, Immune Escape, and Drug Resistance
Abstract
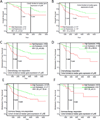
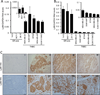
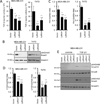
Stem cell antigen Sca-1 is implicated in murine cancer stem cell biology and breast cancer models, but the role of its human homologs Ly6K and Ly6E in breast cancer are not established. Here we report increased expression of Ly6K/E in human breast cancer specimens correlates with poor overall survival, with an additional specific role for Ly6E in poor therapeutic outcomes. Increased expression of Ly6K/E also correlated with increased expression of the immune checkpoint molecules PDL1 and CTLA4, increased tumor-infiltrating T regulatory cells, and decreased natural killer (NK) cell activation. Mechanistically, Ly6K/E was required for TGFβ signaling and proliferation in breast cancer cells, where they contributed to phosphorylation of Smad1/5 and Smad2/3. Furthermore, Ly6K/E promoted cytokine-induced PDL1 expression and activation and binding of NK cells to cancer cells. Finally, we found that Ly6K/E promoted drug resistance and facilitated immune escape in this setting. Overall, our results establish a pivotal role for a Ly6K/E signaling axis involving TGFβ in breast cancer pathophysiology and drug response, and highlight this signaling axis as a compelling realm for therapeutic invention. Cancer Res; 76(11); 3376-86. ©2016 AACR.
©2016 American Association for Cancer Research.
Figures



References
-
- Atena M, Reza AM, Mehran G. Stem Cell Discovery. 04. Scientific Research Publishing; 2014. A Review on the Biology of Cancer Stem Cells; p. 83.
-
- Holmes C, Stanford WL. Concise review: stem cell antigen-1: expression, function, and enigma. Stem Cells. 2007;25:1339–1347. - PubMed
-
- Gumley T, McKenzie I, Sandrin M. Tissue expression, structure and function of the murine Ly-6 family of molecules. Immunology and cell …. 1995 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

