Label-Free Neurosurgical Pathology with Stimulated Raman Imaging
- PMID: 27197198
- PMCID: PMC4911248
- DOI: 10.1158/0008-5472.CAN-16-0270
Label-Free Neurosurgical Pathology with Stimulated Raman Imaging
Abstract
The goal of brain tumor surgery is to maximize tumor removal without injuring critical brain structures. Achieving this goal is challenging as it can be difficult to distinguish tumor from nontumor tissue. While standard histopathology provides information that could assist tumor delineation, it cannot be performed iteratively during surgery as freezing, sectioning, and staining of the tissue require too much time. Stimulated Raman scattering (SRS) microscopy is a powerful label-free chemical imaging technology that enables rapid mapping of lipids and proteins within a fresh specimen. This information can be rendered into pathology-like images. Although this approach has been used to assess the density of glioma cells in murine orthotopic xenografts models and human brain tumors, tissue heterogeneity in clinical brain tumors has not yet been fully evaluated with SRS imaging. Here we profile 41 specimens resected from 12 patients with a range of brain tumors. By evaluating large-scale stimulated Raman imaging data and correlating this data with current clinical gold standard of histopathology for 4,422 fields of view, we capture many essential diagnostic hallmarks for glioma classification. Notably, in fresh tumor samples, we observe additional features, not seen by conventional methods, including extensive lipid droplets within glioma cells, collagen deposition in gliosarcoma, and irregularity and disruption of myelinated fibers in areas infiltrated by oligodendroglioma cells. The data are freely available in a public resource to foster diagnostic training and to permit additional interrogation. Our work establishes the methodology and provides a significant collection of reference images for label-free neurosurgical pathology. Cancer Res; 76(12); 3451-62. ©2016 AACR.
©2016 American Association for Cancer Research.
Figures
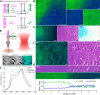

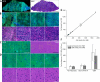
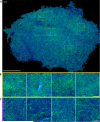
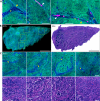

References
-
- McGirt MJ, Chaichana KL, Gathinji M, Attenello FJ, Than K, Olivi A, et al. Independent association of extent of resection with survival in patients with malignant brain astrocytoma. J Neurosurg. 2009;110(1):156–62. - PubMed
-
- Asthagiri AR, Pouratian N, Sherman J, Ahmed G, Shaffrey ME. Advances in brain tumor surgery. Neurol Clin. 2007;25(4):975–1003, viii-ix. - PubMed
-
- Pallud J, Varlet P, Devaux B, Geha S, Badoual M, Deroulers C, et al. Diffuse low-grade oligodendrogliomas extend beyond MRI-defined abnormalities. Neurology. 2010;74(21):1724–31. - PubMed
-
- Kiernan JA. Histological and histochemical methods : theory and practice. Scion; Oxford: 2008.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

