Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast
- PMID: 27197211
- PMCID: PMC4889974
- DOI: 10.1101/gr.204578.116
Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast
Erratum in
-
Corrigendum: Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast.Genome Res. 2016 Jul;26(7):1010-1. doi: 10.1101/gr.210161.116. Genome Res. 2016. PMID: 27371223 Free PMC article. No abstract available.
Abstract
Complex regulation of gene expression in mammals has evolved from simpler eukaryotic systems, yet the mechanistic features of this evolution remain elusive. Here, we compared the transcriptional landscapes of the distantly related budding and fission yeast. We adapted the Precision Run-On sequencing (PRO-seq) approach to map the positions of RNA polymerase active sites genome-wide in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Additionally, we mapped preferred sites of transcription initiation in each organism using PRO-cap. Unexpectedly, we identify a pause in early elongation, specific to S. pombe, that requires the conserved elongation factor subunit Spt4 and resembles promoter-proximal pausing in metazoans. PRO-seq profiles in strains lacking Spt4 reveal globally elevated levels of transcribing RNA Polymerase II (Pol II) within genes in both species. Messenger RNA abundance, however, does not reflect the increases in Pol II density, indicating a global reduction in elongation rate. Together, our results provide the first base-pair resolution map of transcription elongation in S. pombe and identify divergent roles for Spt4 in controlling elongation in budding and fission yeast.
© 2016 Booth et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
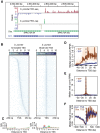


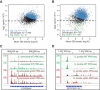
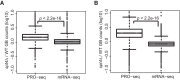
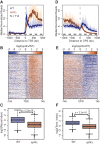
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
