The ribosome in action: Tuning of translational efficiency and protein folding
- PMID: 27198711
- PMCID: PMC4972197
- DOI: 10.1002/pro.2950
The ribosome in action: Tuning of translational efficiency and protein folding
Abstract
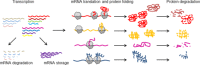
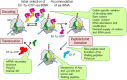
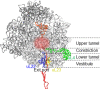
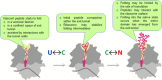
The cellular proteome is shaped by the combined activities of the gene expression and quality control machineries. While transcription plays an undoubtedly important role, in recent years also translation emerged as a key step that defines the composition and quality of the proteome and the functional activity of proteins in the cell. Among the different post-transcriptional control mechanisms, translation initiation and elongation provide multiple checkpoints that can affect translational efficiency. A multitude of specific signals in mRNAs can determine the frequency of translation initiation, choice of the open reading frame, global and local elongation velocities, and the folding of the emerging protein. In addition to specific signatures in the mRNAs, also variations in the global pools of translation components, including ribosomes, tRNAs, mRNAs, and translation factors can alter translational efficiencies. The cellular outcomes of phenomena such as mRNA codon bias are sometimes difficult to understand due to the staggering complexity of covariates that affect codon usage, translation, and protein folding. Here we summarize the experimental evidence on how the ribosome-together with the other components of the translational machinery-can alter translational efficiencies of mRNA at the initiation and elongation stages and how translation velocity affects protein folding. We seek to explain these findings in the context of mechanistic work on the ribosome. The results argue in favour of a new understanding of translation control as a hub that links mRNA homeostasis to production and quality control of proteins in the cell.
Keywords: mRNA rare codons; protein synthesis and folding; ribosome; tRNA; translational pausing.
© 2016 The Protein Society.
Figures


References
-
- Mitarai N, Sneppen K, Pedersen S (2008) Ribosome collisions and translation efficiency: optimization by codon usage and mRNA destabilization. J Mol Biol 382:236–245. - PubMed
-
- Richter JD, Sonenberg N (2005) Regulation of cap‐dependent translation by eIF4E inhibitory proteins. Nature 433:477–480. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

