RNA-Seq methods for transcriptome analysis
- PMID: 27198714
- PMCID: PMC5717752
- DOI: 10.1002/wrna.1364
RNA-Seq methods for transcriptome analysis
Abstract
Deep sequencing has been revolutionizing biology and medicine in recent years, providing single base-level precision for our understanding of nucleic acid sequences in high throughput fashion. Sequencing of RNA, or RNA-Seq, is now a common method to analyze gene expression and to uncover novel RNA species. Aspects of RNA biogenesis and metabolism can be interrogated with specialized methods for cDNA library preparation. In this study, we review current RNA-Seq methods for general analysis of gene expression and several specific applications, including isoform and gene fusion detection, digital gene expression profiling, targeted sequencing and single-cell analysis. In addition, we discuss approaches to examine aspects of RNA in the cell, technical challenges of existing RNA-Seq methods, and future directions. WIREs RNA 2017, 8:e1364. doi: 10.1002/wrna.1364 For further resources related to this article, please visit the WIREs website.
© 2016 Wiley Periodicals, Inc.
Conflict of interest statement
Conflict of interest: Masoud Toloue and Radmila Hrdlickova work at Bioo Scientific, a private corporation.
Figures
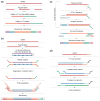
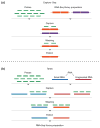
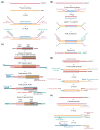

References
-
- Adams MD, Dubnick M, Kerlavage AR, Moreno R, Kelley JM, Utterback TR, Nagle JW, Fields C, Venter JC. Sequence identification of 2,375 human brain genes. Nature. 1992;355:632–634. - PubMed
-
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. - PubMed
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

