Retroviral DNA Integration
- PMID: 27198982
- PMCID: PMC5084067
- DOI: 10.1021/acs.chemrev.6b00125
Retroviral DNA Integration
Abstract
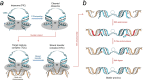
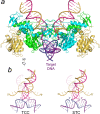
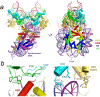
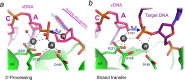
The integration of a DNA copy of the viral RNA genome into host chromatin is the defining step of retroviral replication. This enzymatic process is catalyzed by the virus-encoded integrase protein, which is conserved among retroviruses and LTR-retrotransposons. Retroviral integration proceeds via two integrase activities: 3'-processing of the viral DNA ends, followed by the strand transfer of the processed ends into host cell chromosomal DNA. Herein we review the molecular mechanism of retroviral DNA integration, with an emphasis on reaction chemistries and architectures of the nucleoprotein complexes involved. We additionally discuss the latest advances on anti-integrase drug development for the treatment of AIDS and the utility of integrating retroviral vectors in gene therapy applications.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Ellermann V.; Bang O. Experimentelle Leukämie Bei Hühnern. Zentralbl. Bakteriol. Parasitenkd. Infectionskr. Hyg. Abt. Orig. 1908, 46, 595–609.
-
- Poiesz B. J.; Ruscetti F. W.; Gazdar A. F.; Bunn P. A.; Minna J. D.; Gallo R. C. Detection and Isolation of Type C Retrovirus Particles from Fresh and Cultured Lymphocytes of a Patient with Cutaneous T-Cell Lymphoma. Proc. Natl. Acad. Sci. U. S. A. 1980, 77 (12), 7415–7419. 10.1073/pnas.77.12.7415. - DOI - PMC - PubMed
-
- Barre-Sinoussi F.; Chermann J. C.; Rey F.; Nugeyre M. T.; Chamaret S.; Gruest J.; Dauguet C.; Axler-Blin C.; Vezinet-Brun F.; Rouzioux C.; et al. Isolation of a T-Lymphotropic Retrovirus from a Patient at Risk for Acquired Immune Deficiency Syndrome (AIDS). Science 1983, 220 (4599), 868–871. 10.1126/science.6189183. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

