The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas
- PMID: 27199435
- PMCID: PMC5450629
- DOI: 10.1126/science.aad3018
The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas
Abstract
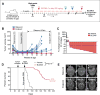
Macrophages accumulate with glioblastoma multiforme (GBM) progression and can be targeted via inhibition of colony-stimulating factor-1 receptor (CSF-1R) to regress high-grade tumors in animal models of this cancer. However, whether and how resistance emerges in response to sustained CSF-1R blockade is unknown. We show that although overall survival is significantly prolonged, tumors recur in >50% of mice. Gliomas reestablish sensitivity to CSF-1R inhibition upon transplantation, indicating that resistance is tumor microenvironment-driven. Phosphatidylinositol 3-kinase (PI3K) pathway activity was elevated in recurrent GBM, driven by macrophage-derived insulin-like growth factor-1 (IGF-1) and tumor cell IGF-1 receptor (IGF-1R). Combining IGF-1R or PI3K blockade with CSF-1R inhibition in recurrent tumors significantly prolonged overall survival. Our findings thus reveal a potential therapeutic approach for treating resistance to CSF-1R inhibitors.
Copyright © 2016, American Association for the Advancement of Science.
Figures


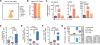




Comment in
-
The Taming of the TAMs.Trends Cell Biol. 2016 Aug;26(8):562-563. doi: 10.1016/j.tcb.2016.06.007. Epub 2016 Jul 6. Trends Cell Biol. 2016. PMID: 27394965
References
-
- Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. - PubMed
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, R. European Organisation for, T. Treatment of Cancer Brain, G. Radiotherapy, G. National Cancer Institute of Canada Clinical Trials Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. - PubMed
-
- Komohara Y, Ohnishi K, Kuratsu J, Takeya M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J Pathol. 2008;216:15–24. - PubMed
-
- Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196:254–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

