Phylogenetic and Structural Analysis of Polyketide Synthases in Aspergilli
- PMID: 27199544
- PMCID: PMC4863872
- DOI: 10.4137/EBO.S32694
Phylogenetic and Structural Analysis of Polyketide Synthases in Aspergilli
Abstract
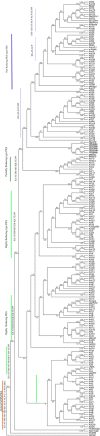
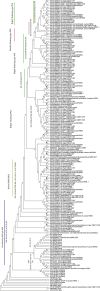
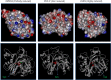
Polyketide synthases (PKSs) of Aspergillus species are multidomain and multifunctional megaenzymes that play an important role in the synthesis of diverse polyketide compounds. Putative PKS protein sequences from Aspergillus species representing medically, agriculturally, and industrially important Aspergillus species were chosen and screened for in silico studies. Six candidate Aspergillus species, Aspergillus fumigatus Af293, Aspergillus flavus NRRL3357, Aspergillus niger CBS 513.88, Aspergillus terreus NIH2624, Aspergillus oryzae RIB40, and Aspergillus clavatus NRRL1, were selected to study the PKS phylogeny. Full-length PKS proteins and only ketosynthase (KS) domain sequence were retrieved for independent phylogenetic analysis from the aforementioned species, and phylogenetic analysis was performed with characterized fungal PKS. This resulted into grouping of Aspergilli PKSs into nonreducing (NR), partially reducing (PR), and highly reducing (HR) PKS enzymes. Eight distinct clades with unique domain arrangements were classified based on homology with functionally characterized PKS enzymes. Conserved motif signatures corresponding to each type of PKS were observed. Three proteins from Protein Data Bank corresponding to NR, PR, and HR type of PKS (XP_002384329.1, XP_753141.2, and XP_001402408.2, respectively) were selected for mapping of conserved motifs on three-dimensional structures of KS domain. Structural variations were found at the active sites on modeled NR, PR, and HR enzymes of Aspergillus. It was observed that the number of iteration cycles was dependent on the size of the cavity in the active site of the PKS enzyme correlating with a type with reducing or NR products, such as pigment, 6MSA, and lovastatin. The current study reports the grouping and classification of PKS proteins of Aspergilli for possible exploration of novel polyketides based on sequence homology; this information can be useful for selection of PKS for polyketide exploration and specific detection of Aspergilli.
Keywords: Aspergillus; ketosynthase; phylogeny; polyketide; polyketide synthases.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

