MYB Transcription Factors in Chinese Pear (Pyrus bretschneideri Rehd.): Genome-Wide Identification, Classification, and Expression Profiling during Fruit Development
- PMID: 27200050
- PMCID: PMC4850919
- DOI: 10.3389/fpls.2016.00577
MYB Transcription Factors in Chinese Pear (Pyrus bretschneideri Rehd.): Genome-Wide Identification, Classification, and Expression Profiling during Fruit Development
Abstract
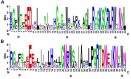
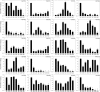
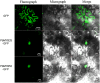
The MYB family is one of the largest families of transcription factors in plants. Although, some MYBs were reported to play roles in secondary metabolism, no comprehensive study of the MYB family in Chinese pear (Pyrus bretschneideri Rehd.) has been reported. In the present study, we performed genome-wide analysis of MYB genes in Chinese pear, designated as PbMYBs, including analyses of their phylogenic relationships, structures, chromosomal locations, promoter regions, GO annotations, and collinearity. A total of 129 PbMYB genes were identified in the pear genome and were divided into 31 subgroups based on phylogenetic analysis. These PbMYBs were unevenly distributed among 16 chromosomes (total of 17 chromosomes). The occurrence of gene duplication events indicated that whole-genome duplication and segmental duplication likely played key roles in expansion of the PbMYB gene family. Ka/Ks analysis suggested that the duplicated PbMYBs mainly experienced purifying selection with restrictive functional divergence after the duplication events. Interspecies microsynteny analysis revealed maximum orthology between pear and peach, followed by plum and strawberry. Subsequently, the expression patterns of 20 PbMYB genes that may be involved in lignin biosynthesis according to their phylogenetic relationships were examined throughout fruit development. Among the 20 genes examined, PbMYB25 and PbMYB52 exhibited expression patterns consistent with the typical variations in the lignin content previously reported. Moreover, sub-cellular localization analysis revealed that two proteins PbMYB25 and PbMYB52 were localized to the nucleus. All together, PbMYB25 and PbMYB52 were inferred to be candidate genes involved in the regulation of lignin biosynthesis during the development of pear fruit. This study provides useful information for further functional analysis of the MYB gene family in pear.
Keywords: MYB transcription factor; expression; gene family; lignin synthesis; pear.
Figures




References
-
- Bailey T. L., Elkan C. (1995). The value of prior knowledge in discovering motifs with MEME. Proc Int. Conf. Intell. Syst. Mol. Biol. 3 21–29. - PubMed
-
- Cai Y., Li G., Nie J., Lin Y., Nie F., Zhang J., et al. (2010). Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Horticult. 125 374–379. 10.1016/j.scienta.2010.04.029 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

