Identification of a Candidate Gene for Panicle Length in Rice (Oryza sativa L.) Via Association and Linkage Analysis
- PMID: 27200064
- PMCID: PMC4853638
- DOI: 10.3389/fpls.2016.00596
Identification of a Candidate Gene for Panicle Length in Rice (Oryza sativa L.) Via Association and Linkage Analysis
Abstract
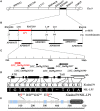
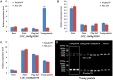
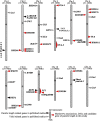
Panicle length (PL) is an important trait for improving panicle architecture and grain yield in rice (Oryza sativa L.). Three populations were used to identify QTLs and candidate genes associated with PL. Four QTLs for PL were detected on chromosomes 4, 6, and 9 through linkage mapping in the recombinant inbred line population derived from a cross between the cultivars Xiushui79 (short panicle) and C-bao (long panicle). Ten SSR markers associated with PL were detected on chromosomes 2, 3, 5, 6, 8, 9, and 10 in the natural population consisting of 540 accessions collected from East and Southeast Asia. A major locus on chromosome 9 with the largest effect was identified via both linkage and association mapping. LONG PANICLE 1 (LP1) locus was delimited to a 90-kb region of the long arm of chromosome 9 through fine mapping using a single segment segregating F2 population. Two single nucleotide polymorphisms (SNPs) leading to amino acid changes were detected in the third and fifth exons of LP1. LP1 encodes a Remorin_C-containing protein of unknown function with homologs in a variety of species. Sequencing analysis of LP1 in two parents and 103 rice accessions indicated that SNP1 is associated with panicle length. The LP1 allele of Xiushui79 leads to reduced panicle length, whereas the allele of C-bao relieves the suppression of panicle length. LP1 and the elite alleles can be used to improve panicle length in rice.
Keywords: association analysis; gene identification; map-based cloning; panicle length; quantitative trait locus; rice.
Figures



Similar articles
-
Quantitative trait locus analysis and fine mapping of the qPL6 locus for panicle length in rice.Theor Appl Genet. 2015 Jun;128(6):1151-61. doi: 10.1007/s00122-015-2496-y. Epub 2015 Mar 28. Theor Appl Genet. 2015. PMID: 25821195
-
Novel pleiotropic loci controlling panicle architecture across environments in japonica rice (Oryza sativa L.).J Genet Genomics. 2010 Aug;37(8):533-44. doi: 10.1016/S1673-8527(09)60073-4. J Genet Genomics. 2010. PMID: 20816386
-
Identification of a Novel QTL for Panicle Length From Wild Rice (Oryza minuta) by Specific Locus Amplified Fragment Sequencing and High Density Genetic Mapping.Front Plant Sci. 2018 Oct 16;9:1492. doi: 10.3389/fpls.2018.01492. eCollection 2018. Front Plant Sci. 2018. PMID: 30459776 Free PMC article.
-
Fine mapping of a major quantitative trait locus, qgnp7(t), controlling grain number per panicle in African rice (Oryza glaberrima S.).Breed Sci. 2018 Dec;68(5):606-613. doi: 10.1270/jsbbs.18084. Epub 2018 Nov 16. Breed Sci. 2018. PMID: 30697122 Free PMC article.
-
A Region on Chromosome 7 Related to Differentiation of Rice (Oryza sativa L.) Between Lowland and Upland Ecotypes.Front Plant Sci. 2020 Jul 27;11:1135. doi: 10.3389/fpls.2020.01135. eCollection 2020. Front Plant Sci. 2020. PMID: 32849696 Free PMC article. Review.
Cited by
-
Putative LysM Effectors Contribute to Fungal Lifestyle.Int J Mol Sci. 2021 Mar 19;22(6):3147. doi: 10.3390/ijms22063147. Int J Mol Sci. 2021. PMID: 33808705 Free PMC article.
-
Detection of QTLs for Plant Height Architecture Traits in Rice (Oryza sativa L.) by Association Mapping and the RSTEP-LRT Method.Plants (Basel). 2022 Apr 6;11(7):999. doi: 10.3390/plants11070999. Plants (Basel). 2022. PMID: 35406978 Free PMC article.
-
Panicle-3D: Efficient Phenotyping Tool for Precise Semantic Segmentation of Rice Panicle Point Cloud.Plant Phenomics. 2021 Dec 23;2021:9838929. doi: 10.34133/2021/9838929. eCollection 2021. Plant Phenomics. 2021. PMID: 35024618 Free PMC article.
-
Assessing and Exploiting Functional Diversity in Germplasm Pools to Enhance Abiotic Stress Adaptation and Yield in Cereals and Food Legumes.Front Plant Sci. 2017 Aug 29;8:1461. doi: 10.3389/fpls.2017.01461. eCollection 2017. Front Plant Sci. 2017. PMID: 28900432 Free PMC article. Review.
-
Deep placement of nitrogen fertilizer improves yield, nitrogen use efficiency and economic returns of transplanted fine rice.PLoS One. 2021 Feb 25;16(2):e0247529. doi: 10.1371/journal.pone.0247529. eCollection 2021. PLoS One. 2021. Retraction in: PLoS One. 2022 Oct 21;17(10):e0275942. doi: 10.1371/journal.pone.0275942. PMID: 33630922 Free PMC article. Retracted.
References
-
- Agrama H. A., Eizenga G. C., Yan W. (2007). Association mapping of yield and its components in rice cultivars. Mol Breeding. 19, 341–356. 10.1007/s11032-006-9066-6 - DOI
-
- Benson J. M., Poland J. A., Benson B. M., Stromberg E. L., Nelson R. J. (2015). Resistance to gray leaf spot of maize: genetic architecture and mechanisms elucidated through nested association mapping and near-isogenic line analysis. PLOS Genet. 11:e1005045. 10.1371/journal.pgen.1005045 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

