Prevalent Exon-Intron Structural Changes in the APETALA1/FRUITFULL, SEPALLATA, AGAMOUS-LIKE6, and FLOWERING LOCUS C MADS-Box Gene Subfamilies Provide New Insights into Their Evolution
- PMID: 27200066
- PMCID: PMC4852290
- DOI: 10.3389/fpls.2016.00598
Prevalent Exon-Intron Structural Changes in the APETALA1/FRUITFULL, SEPALLATA, AGAMOUS-LIKE6, and FLOWERING LOCUS C MADS-Box Gene Subfamilies Provide New Insights into Their Evolution
Abstract
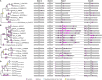
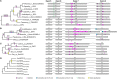
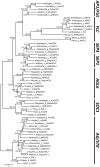
AP1/FUL, SEP, AGL6, and FLC subfamily genes play important roles in flower development. The phylogenetic relationships among them, however, have been controversial, which impedes our understanding of the origin and functional divergence of these genes. One possible reason for the controversy may be the problems caused by changes in the exon-intron structure of genes, which, according to recent studies, may generate non-homologous sites and hamper the homology-based sequence alignment. In this study, we first performed exon-by-exon alignments of these and three outgroup subfamilies (SOC1, AG, and STK). Phylogenetic trees reconstructed based on these matrices show improved resolution and better congruence with species phylogeny. In the context of these phylogenies, we traced evolutionary changes of exon-intron structures in each subfamily. We found that structural changes have occurred frequently following gene duplication and speciation events. Notably, exons 7 and 8 (if present) suffered more structural changes than others. With the knowledge of exon-intron structural changes, we generated more reasonable alignments containing all the focal subfamilies. The resulting trees showed that the SEP subfamily is sister to the monophyletic group formed by AP1/FUL and FLC subfamily genes and that the AGL6 subfamily forms a sister group to the three abovementioned subfamilies. Based on this topology, we inferred the evolutionary history of exon-intron structural changes among different subfamilies. Particularly, we found that the eighth exon originated before the divergence of AP1/FUL, FLC, SEP, and AGL6 subfamilies and degenerated in the ancestral FLC-like gene. These results provide new insights into the origin and evolution of the AP1/FUL, FLC, SEP, and AGL6 subfamilies.
Keywords: AGAMOUS-LIKE6; APETALA1/FRUITFULL; FLOWERING LOCUS C; SEPALLATA; exon-intron structural change.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

