Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape
- PMID: 27203113
- PMCID: PMC4907330
- DOI: 10.1016/j.cell.2016.04.038
Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape
Erratum in
-
Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape.Cell. 2016 Sep 8;166(6):1598. doi: 10.1016/j.cell.2016.08.063. Epub 2016 Sep 8. Cell. 2016. PMID: 27610578 No abstract available.
Abstract
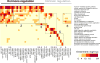
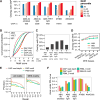
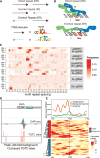
The cistrome is the complete set of transcription factor (TF) binding sites (cis-elements) in an organism, while an epicistrome incorporates tissue-specific DNA chemical modifications and TF-specific chemical sensitivities into these binding profiles. Robust methods to construct comprehensive cistrome and epicistrome maps are critical for elucidating complex transcriptional networks that underlie growth, behavior, and disease. Here, we describe DNA affinity purification sequencing (DAP-seq), a high-throughput TF binding site discovery method that interrogates genomic DNA with in-vitro-expressed TFs. Using DAP-seq, we defined the Arabidopsis cistrome by resolving motifs and peaks for 529 TFs. Because genomic DNA used in DAP-seq retains 5-methylcytosines, we determined that >75% (248/327) of Arabidopsis TFs surveyed were methylation sensitive, a property that strongly impacts the epicistrome landscape. DAP-seq datasets also yielded insight into the biology and binding site architecture of numerous TFs, demonstrating the value of DAP-seq for cost-effective cistromic and epicistromic annotation in any organism.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




References
-
- Boer DR, Freire-Rios A, van den Berg WAM, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014;156:577–589. - PubMed
-
- Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008;18:756–767. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

