Recognition of Lys48-Linked Di-ubiquitin and Deubiquitinating Activities of the SARS Coronavirus Papain-like Protease
- PMID: 27203180
- PMCID: PMC4875570
- DOI: 10.1016/j.molcel.2016.04.016
Recognition of Lys48-Linked Di-ubiquitin and Deubiquitinating Activities of the SARS Coronavirus Papain-like Protease
Abstract
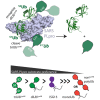
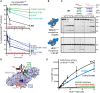
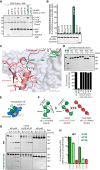
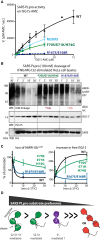
Deubiquitinating enzymes (DUBs) recognize and cleave linkage-specific polyubiquitin (polyUb) chains, but mechanisms underlying specificity remain elusive in many cases. The severe acute respiratory syndrome (SARS) coronavirus papain-like protease (PLpro) is a DUB that cleaves ISG15, a two-domain Ub-like protein, and Lys48-linked polyUb chains, releasing diUb(Lys48) products. To elucidate this specificity, we report the 2.85 Å crystal structure of SARS PLpro bound to a diUb(Lys48) activity-based probe. SARS PLpro binds diUb(Lys48) in an extended conformation via two contact sites, S1 and S2, which are proximal and distal to the active site, respectively. We show that specificity for polyUb(Lys48) chains is predicated on contacts in the S2 site and enhanced by an S1-S1' preference for a Lys48 linkage across the active site. In contrast, ISG15 specificity is dominated by contacts in the S1 site. Determinants revealed for polyUb(Lys48) specificity should prove useful in understanding PLpro deubiquitinating activities in coronavirus infections.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


References
-
- Bailey-Elkin B.A., Knaap R.C., Johnson G.G., Dalebout T.J., Ninaber D.K., van Kasteren P.B., Bredenbeek P.J., Snijder E.J., Kikkert M., Mark B.L. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289:34667–34682. - PMC - PubMed
-
- Berger A., Schechter I. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1970;257:249–264. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

