Antimicrobial Functions of Lactoferrin Promote Genetic Conflicts in Ancient Primates and Modern Humans
- PMID: 27203426
- PMCID: PMC4874600
- DOI: 10.1371/journal.pgen.1006063
Antimicrobial Functions of Lactoferrin Promote Genetic Conflicts in Ancient Primates and Modern Humans
Abstract
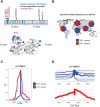
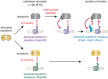
Lactoferrin is a multifunctional mammalian immunity protein that limits microbial growth through sequestration of nutrient iron. Additionally, lactoferrin possesses cationic protein domains that directly bind and inhibit diverse microbes. The implications for these dual functions on lactoferrin evolution and genetic conflicts with microbes remain unclear. Here we show that lactoferrin has been subject to recurrent episodes of positive selection during primate divergence predominately at antimicrobial peptide surfaces consistent with long-term antagonism by bacteria. An abundant lactoferrin polymorphism in human populations and Neanderthals also exhibits signatures of positive selection across primates, linking ancient host-microbe conflicts to modern human genetic variation. Rapidly evolving sites in lactoferrin further correspond to molecular interfaces with opportunistic bacterial pathogens causing meningitis, pneumonia, and sepsis. Because microbes actively target lactoferrin to acquire iron, we propose that the emergence of antimicrobial activity provided a pivotal mechanism of adaptation sparking evolutionary conflicts via acquisition of new protein functions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Molecular Basis for the Evolution of Species-Specific Hemoglobin Capture by Staphylococcus aureus.mBio. 2018 Nov 20;9(6):e01524-18. doi: 10.1128/mBio.01524-18. mBio. 2018. PMID: 30459189 Free PMC article.
-
Bacterial receptors for host transferrin and lactoferrin: molecular mechanisms and role in host-microbe interactions.Future Microbiol. 2013 Dec;8(12):1575-85. doi: 10.2217/fmb.13.125. Future Microbiol. 2013. PMID: 24266357 Review.
-
Rapid Evolution of Primate Type 2 Immune Response Factors Linked to Asthma Susceptibility.Genome Biol Evol. 2017 Jun 1;9(6):1757-1765. doi: 10.1093/gbe/evx120. Genome Biol Evol. 2017. PMID: 28854632 Free PMC article.
-
The specificity of protection against cationic antimicrobial peptides by lactoferrin binding protein B.Biometals. 2014 Oct;27(5):923-33. doi: 10.1007/s10534-014-9767-y. Epub 2014 Jul 20. Biometals. 2014. PMID: 25038734
-
Lactoferrin--a multifunctional protein with antimicrobial properties.Mol Immunol. 2003 Nov;40(7):395-405. doi: 10.1016/s0161-5890(03)00152-4. Mol Immunol. 2003. PMID: 14568385 Review.
Cited by
-
Battlefronts of evolutionary conflict between bacteria and animal hosts.PLoS Pathog. 2020 Sep 17;16(9):e1008797. doi: 10.1371/journal.ppat.1008797. eCollection 2020 Sep. PLoS Pathog. 2020. PMID: 32941529 Free PMC article. No abstract available.
-
Molecular de-extinction of ancient antimicrobial peptides enabled by machine learning.Cell Host Microbe. 2023 Aug 9;31(8):1260-1274.e6. doi: 10.1016/j.chom.2023.07.001. Epub 2023 Jul 28. Cell Host Microbe. 2023. PMID: 37516110 Free PMC article.
-
Lactoferrin is a dynamic protein in human melioidosis and is a TLR4-dependent driver of TNF-α release in Burkholderia thailandensis infection in vitro.PLoS Negl Trop Dis. 2020 Aug 7;14(8):e0008495. doi: 10.1371/journal.pntd.0008495. eCollection 2020 Aug. PLoS Negl Trop Dis. 2020. PMID: 32764765 Free PMC article.
-
Lactoferrin in a Context of Inflammation-Induced Pathology.Front Immunol. 2017 Nov 6;8:1438. doi: 10.3389/fimmu.2017.01438. eCollection 2017. Front Immunol. 2017. PMID: 29163511 Free PMC article. Review.
-
Oral-Delivery Lactococcus lactis expressing cherry fusion lactoferrin peptides against infection of avian pathogenic Escherichia coli in chickens.Poult Sci. 2025 Jan;104(1):104637. doi: 10.1016/j.psj.2024.104637. Epub 2024 Dec 4. Poult Sci. 2025. PMID: 39662258 Free PMC article.
References
-
- Haldane J. Disease and evolution. La Ricerca Scientifica Supplemento. 1949;: 1–11.
-
- Van Valen L. A new evolutionary law. Evol Theory. 1973;1: 1–30.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

