High Efficiency CRISPR/Cas9-mediated Gene Editing in Primary Human T-cells Using Mutant Adenoviral E4orf6/E1b55k "Helper" Proteins
- PMID: 27203437
- PMCID: PMC5113096
- DOI: 10.1038/mt.2016.105
High Efficiency CRISPR/Cas9-mediated Gene Editing in Primary Human T-cells Using Mutant Adenoviral E4orf6/E1b55k "Helper" Proteins
Abstract
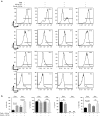
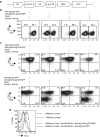
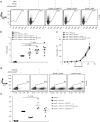
Many future therapeutic applications of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 and related RNA-guided nucleases are likely to require their use to promote gene targeting, thus necessitating development of methods that provide for delivery of three components-Cas9, guide RNAs and recombination templates-to primary cells rendered proficient for homology-directed repair. Here, we demonstrate an electroporation/transduction codelivery method that utilizes mRNA to express both Cas9 and mutant adenoviral E4orf6 and E1b55k helper proteins in association with adeno-associated virus (AAV) vectors expressing guide RNAs and recombination templates. By transiently enhancing target cell permissiveness to AAV transduction and gene editing efficiency, this novel approach promotes efficient gene disruption and/or gene targeting at multiple loci in primary human T-cells, illustrating its broad potential for application in translational gene editing.
Figures


References
-
- Gori, JL, Hsu, PD, Maeder, ML, Shen, S, Welstead, GG and Bumcrot, D (2015). Delivery and specificity of CRISPR-Cas9 genome editing technologies for human gene therapy. Hum Gene Ther 26: 443–451. - PubMed
-
- Jiang, W and Marraffini, LA (2015). CRISPR-Cas: New tools for genetic manipulations from bacterial immunity systems. Annu Rev Microbiol 69: 209–228. - PubMed
-
- Xu, J, Ren, X, Sun, J, Wang, X, Qiao, HH, Xu, BW et al. (2015). A toolkit of CRISPR-based genome editing systems in Drosophila. J Genet Genomics 42: 141–149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

