Reactome from a WikiPathways Perspective
- PMID: 27203685
- PMCID: PMC4874630
- DOI: 10.1371/journal.pcbi.1004941
Reactome from a WikiPathways Perspective
Abstract
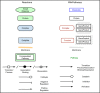
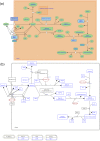
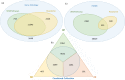
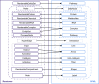
Reactome and WikiPathways are two of the most popular freely available databases for biological pathways. Reactome pathways are centrally curated with periodic input from selected domain experts. WikiPathways is a community-based platform where pathways are created and continually curated by any interested party. The nascent collaboration between WikiPathways and Reactome illustrates the mutual benefits of combining these two approaches. We created a format converter that converts Reactome pathways to the GPML format used in WikiPathways. In addition, we developed the ComplexViz plugin for PathVisio which simplifies looking up complex components. The plugin can also score the complexes on a pathway based on a user defined criterion. This score can then be visualized on the complex nodes using the visualization options provided by the plugin. Using the merged collection of curated and converted Reactome pathways, we demonstrate improved pathway coverage of relevant biological processes for the analysis of a previously described polycystic ovary syndrome gene expression dataset. Additionally, this conversion allows researchers to visualize their data on Reactome pathways using PathVisio's advanced data visualization functionalities. WikiPathways benefits from the dedicated focus and attention provided to the content converted from Reactome and the wealth of semantic information about interactions. Reactome in turn benefits from the continuous community curation available on WikiPathways. The research community at large benefits from the availability of a larger set of pathways for analysis in PathVisio and Cytoscape. The pathway statistics results obtained from PathVisio are significantly better when using a larger set of candidate pathways for analysis. The conversion serves as a general model for integration of multiple pathway resources developed using different approaches.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

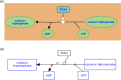
References
-
- PathGuide the pathway resource list [18-07-2015]. Available from: http://pathguide.org/.
-
- WikiPathways Curation Protocol. Available from: http://wikipathways.org/index.php/Help:Curation_Protocol.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

