Premature polyadenylation of MAGI3 produces a dominantly-acting oncogene in human breast cancer
- PMID: 27205883
- PMCID: PMC4905742
- DOI: 10.7554/eLife.14730
Premature polyadenylation of MAGI3 produces a dominantly-acting oncogene in human breast cancer
Abstract
Genetic mutation, chromosomal rearrangement and copy number amplification are common mechanisms responsible for generating gain-of-function, cancer-causing alterations. Here we report a new mechanism by which premature cleavage and polyadenylation (pPA) of RNA can produce an oncogenic protein. We identify a pPA event at a cryptic intronic poly(A) signal in MAGI3, occurring in the absence of local exonic and intronic mutations. The altered mRNA isoform, called MAGI3(pPA), produces a truncated protein that acts in a dominant-negative manner to prevent full-length MAGI3 from interacting with the YAP oncoprotein, thereby relieving YAP inhibition and promoting malignant transformation of human mammary epithelial cells. We additionally find evidence for recurrent expression of MAGI3(pPA) in primary human breast tumors but not in tumor-adjacent normal tissues. Our results provide an example of how pPA contributes to cancer by generating a truncated mRNA isoform that encodes an oncogenic, gain-of-function protein.
Keywords: Hippo pathway; MAGI3; YAP; breast cancer; chromosomes; cleavage and polyadenylation; cryptic poly(A) signal; genes; human; human biology; medicine; mouse.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
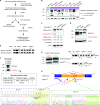





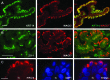
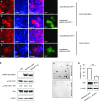


References
-
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, Cortes ML, Fernandez-Lopez JC, Peng S, Ardlie KG, Auclair D, Bautista-Piña V, Duke F, Francis J, Jung J, Maffuz-Aziz A, Onofrio RC, Parkin M, Pho NH, Quintanar-Jurado V, Ramos AH, Rebollar-Vega R, Rodriguez-Cuevas S, Romero-Cordoba SL, Schumacher SE, Stransky N, Thompson KM, Uribe-Figueroa L, Baselga J, Beroukhim R, Polyak K, Sgroi DC, Richardson AL, Jimenez-Sanchez G, Lander ES, Gabriel SB, Garraway LA, Golub TR, Melendez-Zajgla J, Toker A, Getz G, Hidalgo-Miranda A, Meyerson M. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. - DOI - PMC - PubMed
-
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

