Identification and functional characterization of the sulfate transporter gene GmSULTR1;2b in soybean
- PMID: 27206527
- PMCID: PMC4874011
- DOI: 10.1186/s12864-016-2705-3
Identification and functional characterization of the sulfate transporter gene GmSULTR1;2b in soybean
Abstract
Background: Soybean is a major source of oil and protein in the human diet and in animal feed. However, as soybean is deficient in sulfur-containing amino acids, its nutritional value is limited. Increasing sulfate assimilation and utilization efficiency is a valuable approach to augment the concentration of sulfur-containing amino acids in soybean seeds, and sulfate transporters play important roles in both sulfate uptake and translocation within plants.
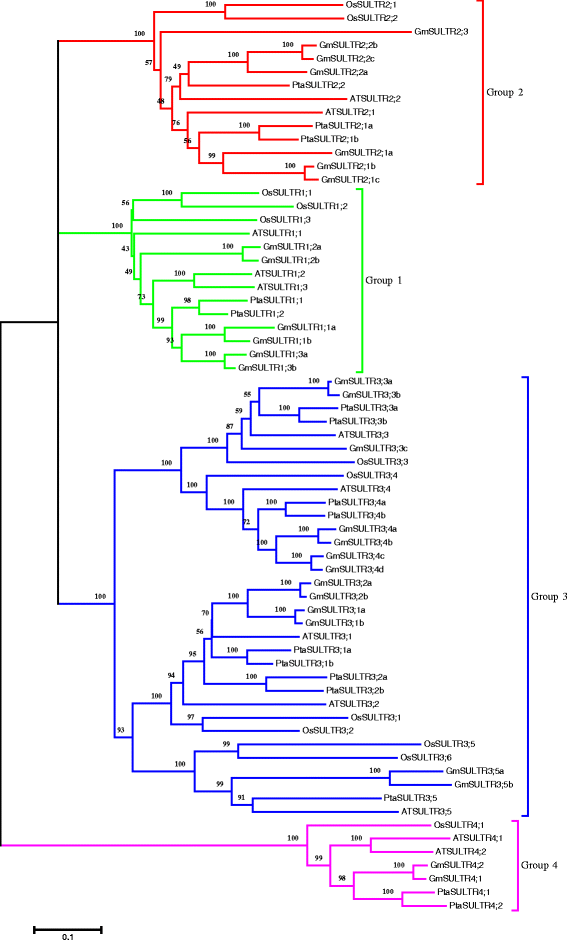
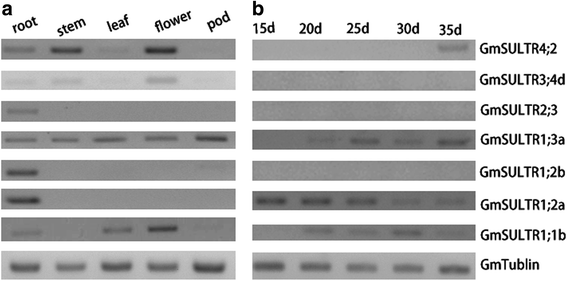
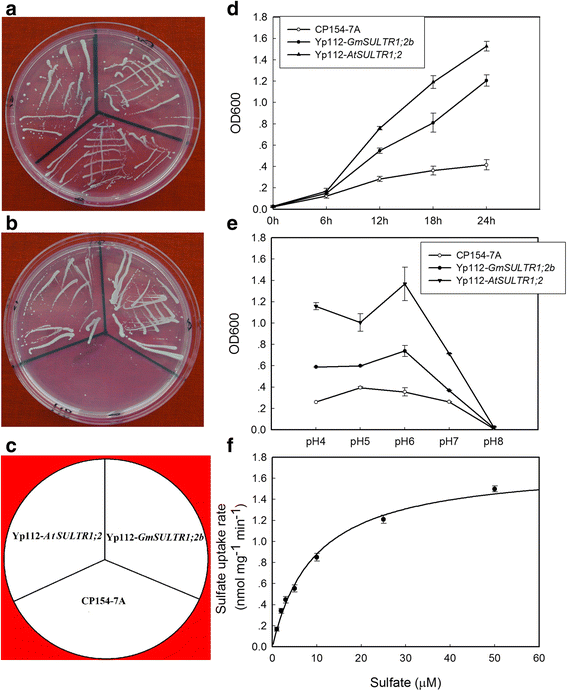
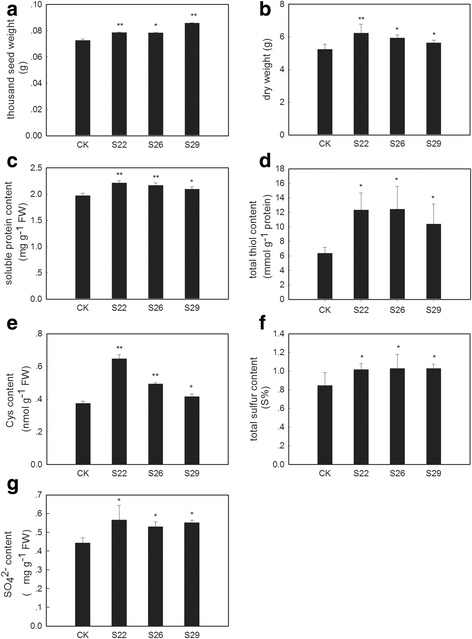
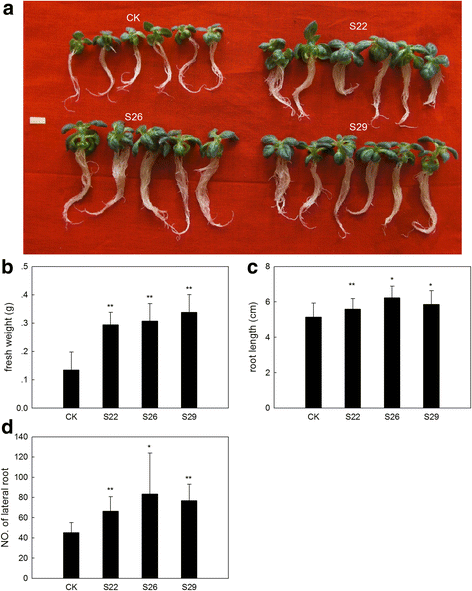
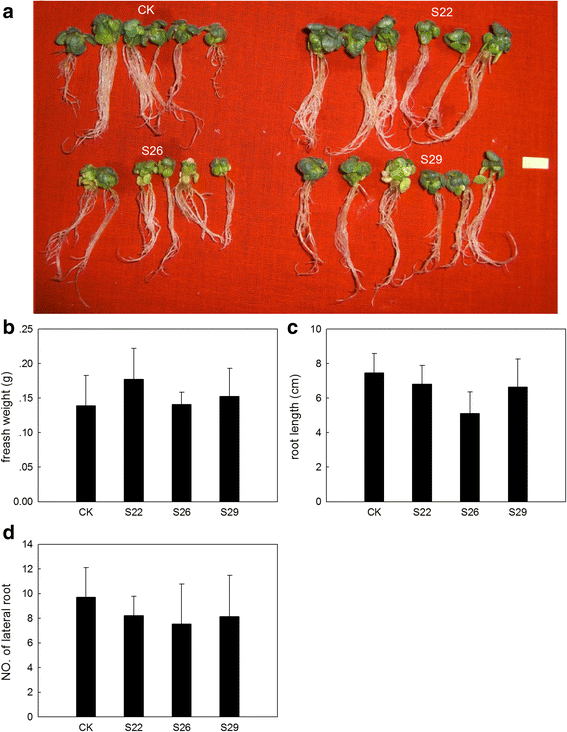
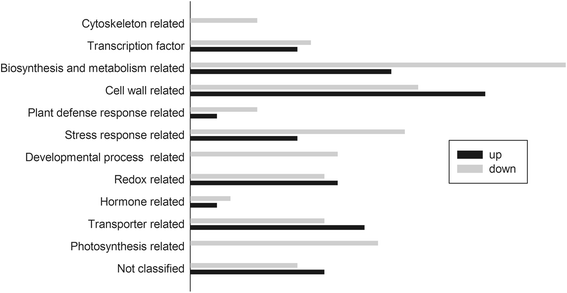
Results: In this study, we isolated and characterized a soybean sulfate transporter gene: GmSULTR1;2b. The gene was found to be specifically expressed in root tissues and induced by low-sulfur stress. In addition, GmSULTR1;2b expression in yeast could complement deficiency in the sulfate transporter genes SUL1 and SUL2. Under +S conditions, GmSULTR1;2b-overexpressing tobacco plants accumulated higher levels of organic matter and exhibited enhanced biomass and seed weight compared to control plants. Under -S conditions, acclimation of GmSULTR1;2b-overexpressing plants was much better than control plants. GmSULTR1;2b-overexpressing tobacco seedlings showed better tolerance to drought stress than the control plants, but no significant difference was observed under salt stress. Transcriptome analysis revealed 515 genes with at least a 2-fold change in expression in the leaves of tobacco plants overexpressing GmSULTR1;2b. Of these, 227 gene annotations were classified into 12 functional categories associated with 123 relevant pathways, including biosynthesis and metabolism-related proteins, stress-related proteins, and transporters.
Conclusions: The findings reported here indicate that the increased biomass and seed yield observed in transgenic tobacco plants could have resulted from greater nutrient uptake and transport capability as well as enhanced development and accumulation of organic matter. Taken together, our results indicate that GmSULTR1;2b plays an important role in sulfur uptake and could alter the sulfur status of plants. Our study suggests that overexpressing GmSULTR1;2b may enhance plant yield under +S conditions, reduce plant production loss under -S conditions, and improve plant tolerance to sulfur deficiency stress.
Keywords: GmSULTR1;2b; Low-sulfur stress; Soybean; Sulfate transporter; Sulfate uptake.
Figures




References
-
- Marsolaisa F, Pajaka A, Yin FQ, Taylora M, Gabriela M, Merino DM, et al. Proteomic analysis of common bean seed with storage protein deficiency reveals up-regulation of sulfur-rich proteins and starch and raffinose metabolic enzymes, and down-regulation of the secretory pathway. J Proteomics. 2010;73:1587–600. doi: 10.1016/j.jprot.2010.03.013. - DOI - PubMed
-
- Scherer HW. Sulphur in crop production–invited paper. Europ J Agron. 2001;14:81–111. doi: 10.1016/S1161-0301(00)00082-4. - DOI
-
- Bao R, Zheng S. Progress on soil sulfur fertility and crop sulfur nutrition. J Heilongjiang Bayi Agricultural University. 2006;18:37–40.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

