Evolution of a global regulator: Lrp in four orders of γ-Proteobacteria
- PMID: 27206730
- PMCID: PMC4875751
- DOI: 10.1186/s12862-016-0685-1
Evolution of a global regulator: Lrp in four orders of γ-Proteobacteria
Abstract
Background: Bacterial global regulators each regulate the expression of several hundred genes. In Escherichia coli, the top seven global regulators together control over half of all genes. Leucine-responsive regulatory protein (Lrp) is one of these top seven global regulators. Lrp orthologs are very widely distributed, among both Bacteria and Archaea. Surprisingly, even within the phylum γ-Proteobacteria (which includes E. coli), Lrp is a global regulator in some orders and a local regulator in others. This raises questions about the evolution of Lrp and, more broadly, of global regulators.
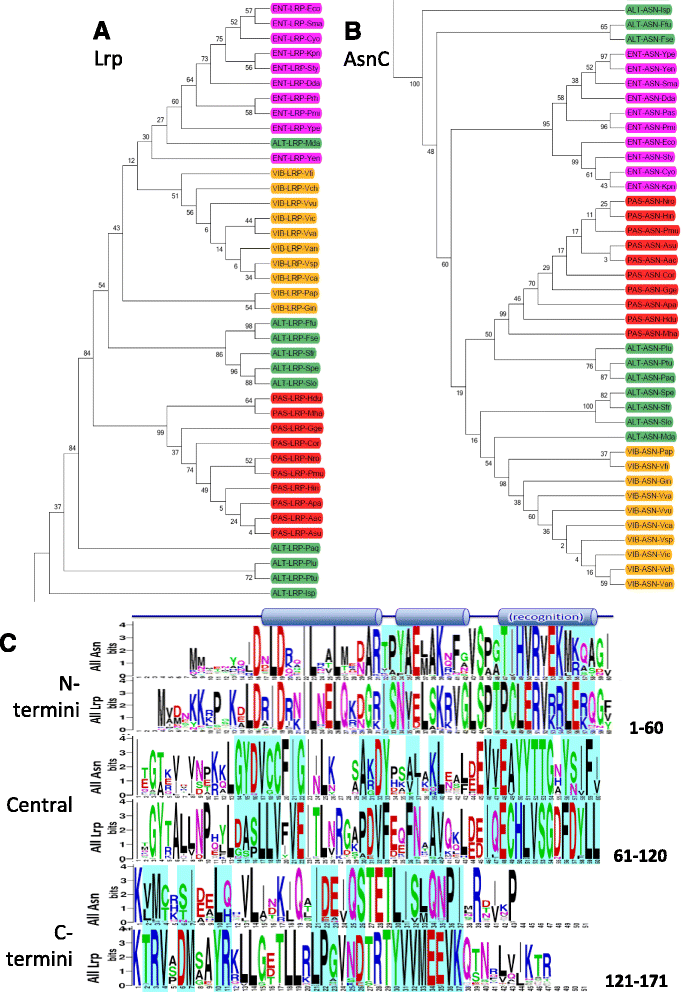
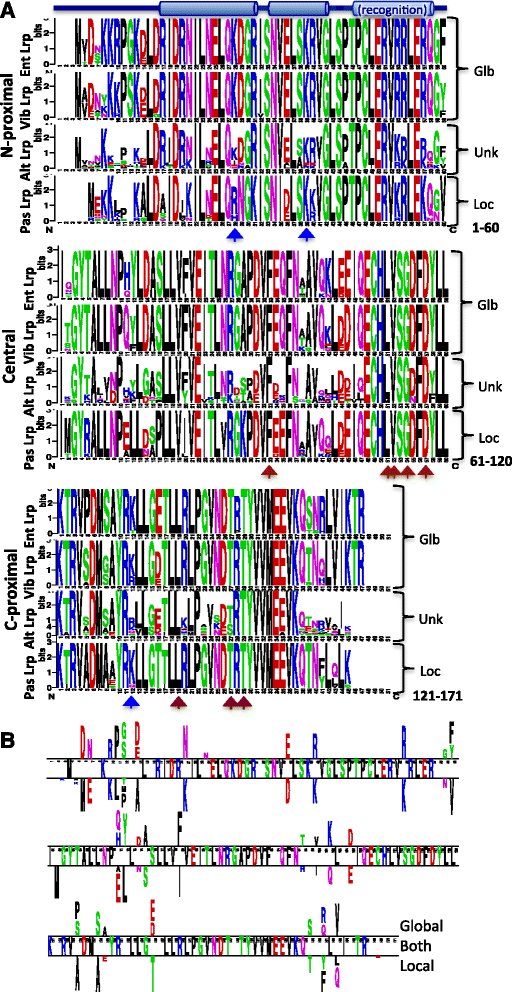
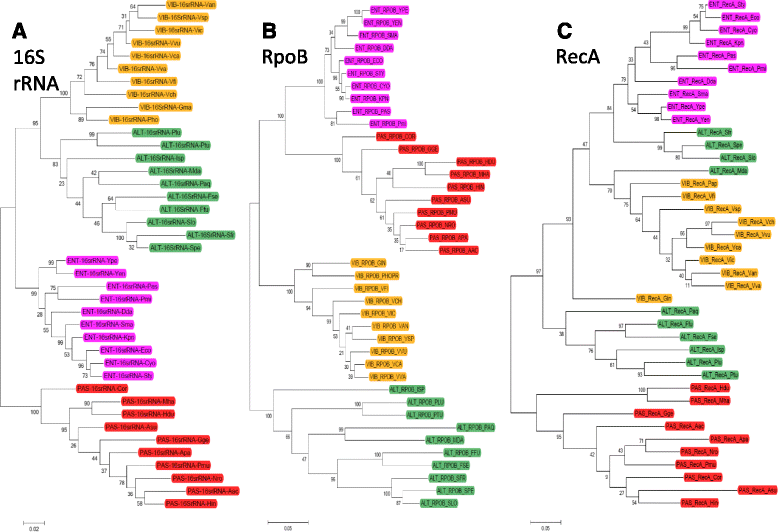
Results: We examined Lrp sequences from four bacterial orders of the γ-Proteobacteria using phylogenetic and Logo analyses. The orders studied were Enterobacteriales and Vibrionales, in which Lrp plays a global role in tested species; Pasteurellales, in which Lrp is a local regulator in the tested species; and Alteromonadales, an order closely related to the other three but in which Lrp has not yet been studied. For comparison, we analyzed the Lrp paralog AsnC, which in all tested cases is a local regulator. The Lrp and AsnC phylogenetic clusters each divided, as expected, into subclusters representing the Enterobacteriales, Vibrionales, and Pasteuralles. However the Alteromonadales did not yield coherent clusters for either Lrp or AsnC. Logo analysis revealed signatures associated with globally- vs. locally- acting Lrp orthologs, providing testable hypotheses for which portions of Lrp are responsible for a global vs. local role. These candidate regions include both ends of the Lrp polypeptide but not, interestingly, the highly-conserved helix-turn-helix motif responsible for DNA sequence specificity.
Conclusions: Lrp and AsnC have conserved sequence signatures that allow their unambiguous annotation, at least in γ-Proteobacteria. Among Lrp orthologs, specific residues correlated with global vs. local regulatory roles, and can now be tested to determine which are functionally relevant and which simply reflect divergence. In the Alteromonadales, it appears that there are different subgroups of Lrp orthologs, one of which may act globally while the other may act locally. These results suggest experiments to improve our understanding of the evolution of bacterial global regulators.
Keywords: Alteromonadales; Enterobacteriales; Pasteurellales; Phylogenomics; Transcription factors; Vibrionales.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

