Engineering a robust DNA split proximity circuit with minimized circuit leakage
- PMID: 27207880
- PMCID: PMC5001597
- DOI: 10.1093/nar/gkw447
Engineering a robust DNA split proximity circuit with minimized circuit leakage
Abstract
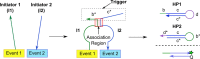
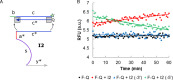
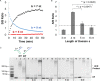
DNA circuit is a versatile and highly-programmable toolbox which can potentially be used for the autonomous sensing of dynamic events, such as biomolecular interactions. However, the experimental implementation of in silico circuit designs has been hindered by the problem of circuit leakage. Here, we systematically analyzed the sources and characteristics of various types of leakage in a split proximity circuit which was engineered to spatially probe for target sites held within close proximity. Direct evidence that 3'-truncated oligonucleotides were the major impurity contributing to circuit leakage was presented. More importantly, a unique strategy of translocating a single nucleotide between domains, termed 'inter-domain bridging', was introduced to eliminate toehold-independent leakages while enhancing the strand displacement kinetics across a three-way junction. We also analyzed the dynamics of intermediate complexes involved in the circuit computation in order to define the working range of domain lengths for the reporter toehold and association region respectively. The final circuit design was successfully implemented on a model streptavidin-biotin system and demonstrated to be robust against both circuit leakage and biological interferences. We anticipate that this simple signal transduction strategy can be used to probe for diverse biomolecular interactions when used in conjunction with specific target recognition moieties.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Engineering high-performance hairpin stacking circuits for logic gate operation and highly sensitive biosensing assay of microRNA.Analyst. 2017 Dec 4;142(24):4834-4842. doi: 10.1039/c7an01624g. Analyst. 2017. PMID: 29160870
-
A Spatially Controlled Proximity Split Tweezer Switch for Enhanced DNA Circuit Construction and Multifunctional Transduction.Small. 2024 May;20(18):e2307421. doi: 10.1002/smll.202307421. Epub 2023 Dec 10. Small. 2024. PMID: 38072808
-
Kinetics of Proximity-Induced Intramolecular DNA Strand Displacement.Anal Chem. 2016 Aug 16;88(16):8152-7. doi: 10.1021/acs.analchem.6b01900. Epub 2016 Aug 4. Anal Chem. 2016. PMID: 27454138
-
Programmable DNA scaffolds for spatially-ordered protein assembly.Nanoscale. 2016 Feb 28;8(8):4436-46. doi: 10.1039/c5nr08685j. Nanoscale. 2016. PMID: 26852879 Review.
-
Streptavidin-biotin technology: improvements and innovations in chemical and biological applications.Appl Microbiol Biotechnol. 2013 Nov;97(21):9343-53. doi: 10.1007/s00253-013-5232-z. Epub 2013 Sep 22. Appl Microbiol Biotechnol. 2013. PMID: 24057405 Review.
Cited by
-
Constructing Controllable Logic Circuits Based on DNAzyme Activity.Molecules. 2019 Nov 15;24(22):4134. doi: 10.3390/molecules24224134. Molecules. 2019. PMID: 31731630 Free PMC article.
-
Orthogonal regulation of DNA nanostructure self-assembly and disassembly using antibodies.Nat Commun. 2019 Dec 3;10(1):5509. doi: 10.1038/s41467-019-13104-6. Nat Commun. 2019. PMID: 31796740 Free PMC article.
-
Designed and Evolved Nucleic Acid Nanotechnology: Contrast and Complementarity.Bioconjug Chem. 2019 Jan 16;30(1):2-12. doi: 10.1021/acs.bioconjchem.8b00810. Epub 2019 Jan 3. Bioconjug Chem. 2019. PMID: 30561987 Free PMC article. Review.
-
Establishment of a universal and rational gene detection strategy through three-way junction-based remote transduction.Chem Sci. 2017 Nov 27;9(3):760-769. doi: 10.1039/c7sc03190d. eCollection 2018 Jan 21. Chem Sci. 2017. PMID: 29629146 Free PMC article.
-
Dynamically elongated associative toehold for tuning DNA circuit kinetics and thermodynamics.Nucleic Acids Res. 2021 May 7;49(8):4258-4265. doi: 10.1093/nar/gkab212. Nucleic Acids Res. 2021. PMID: 33849054 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

