Molecular Epidemiology of Campylobacter coli Strains Isolated from Different Sources in New Zealand between 2005 and 2014
- PMID: 27208097
- PMCID: PMC4959208
- DOI: 10.1128/AEM.00934-16
Molecular Epidemiology of Campylobacter coli Strains Isolated from Different Sources in New Zealand between 2005 and 2014
Abstract
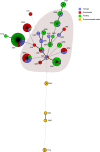
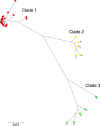
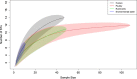
Campylobacteriosis is one of the most important foodborne diseases worldwide and a significant health burden in New Zealand. Campylobacter jejuni is the predominant species worldwide, accounting for approximately 90% of human cases, followed by Campylobacter coli Most studies in New Zealand have focused on C. jejuni; hence, the impact of C. coli strains on human health is not well understood. The aim of this study was to genotype C. coli isolates collected in the Manawatu region of New Zealand from clinical cases, fresh poultry meat, ruminant feces, and environmental water sources, between 2005 and 2014, to study their population structure and estimate the contribution of each source to the burden of human disease. Campylobacter isolates were identified by PCR and typed by multilocus sequence typing. C. coli accounted for 2.9% (n = 47/1,601) of Campylobacter isolates from human clinical cases, 9.6% (n = 108/1,123) from poultry, 13.4% (n = 49/364) from ruminants, and 6.4% (n = 11/171) from water. Molecular subtyping revealed 27 different sequence types (STs), of which 18 belonged to clonal complex ST-828. ST-1581 was the most prevalent C. coli sequence type isolated from both human cases (n = 12/47) and poultry (n = 44/110). When classified using cladistics, all sequence types belonged to clade 1 except ST-7774, which belonged to clade 2. ST-854, ST-1590, and ST-4009 were isolated only from human cases and fresh poultry, while ST-3232 was isolated only from human cases and ruminant sources. Modeling indicated ruminants and poultry as the main sources of C. coli human infection.
Importance: We performed a molecular epidemiological study of Campylobacter coli infection in New Zealand, one of few such studies globally. This study analyzed the population genetic structure of the bacterium and included a probabilistic source attribution model covering different animal and water sources. The results are discussed in a global context.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Shifts in the Molecular Epidemiology of Campylobacter jejuni Infections in a Sentinel Region of New Zealand following Implementation of Food Safety Interventions by the Poultry Industry.Appl Environ Microbiol. 2020 Feb 18;86(5):e01753-19. doi: 10.1128/AEM.01753-19. Print 2020 Feb 18. Appl Environ Microbiol. 2020. PMID: 31862724 Free PMC article.
-
Campylobacter jejuni Strains Associated with Wild Birds and Those Causing Human Disease in Six High-Use Recreational Waterways in New Zealand.Appl Environ Microbiol. 2019 Nov 27;85(24):e01228-19. doi: 10.1128/AEM.01228-19. Print 2019 Dec 15. Appl Environ Microbiol. 2019. PMID: 31562175 Free PMC article.
-
Multilocus sequence typing of Campylobacter jejuni and Campylobacter coli isolates from poultry, cattle and humans in Nigeria.J Appl Microbiol. 2016 Aug;121(2):561-8. doi: 10.1111/jam.13185. J Appl Microbiol. 2016. PMID: 27206561
-
Phenotypic and genotypic methods for typing Campylobacter jejuni and Campylobacter coli in poultry.Poult Sci. 2012 Jan;91(1):255-64. doi: 10.3382/ps.2011-01414. Poult Sci. 2012. PMID: 22184452 Review.
-
A systematic review of source attribution of human campylobacteriosis using multilocus sequence typing.Euro Surveill. 2019 Oct;24(43):1800696. doi: 10.2807/1560-7917.ES.2019.24.43.1800696. Euro Surveill. 2019. PMID: 31662159 Free PMC article.
Cited by
-
Genome-Wide Identification of Host-Segregating Single-Nucleotide Polymorphisms for Source Attribution of Clinical Campylobacter coli Isolates.Appl Environ Microbiol. 2020 Nov 24;86(24):e01787-20. doi: 10.1128/AEM.01787-20. Print 2020 Nov 24. Appl Environ Microbiol. 2020. PMID: 33036986 Free PMC article.
-
The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017.EFSA J. 2019 Feb 27;17(2):e05598. doi: 10.2903/j.efsa.2019.5598. eCollection 2019 Feb. EFSA J. 2019. PMID: 32626224 Free PMC article.
-
Genome sequence of a multidrug-resistant Campylobacter coli strain isolated from a newborn with severe diarrhea in Lebanon.Folia Microbiol (Praha). 2022 Apr;67(2):319-328. doi: 10.1007/s12223-021-00921-w. Epub 2022 Jan 8. Folia Microbiol (Praha). 2022. PMID: 34997523
-
Molecular Identification of Multidrug-Resistant Campylobacter Species From Diarrheal Patients and Poultry Meat in Shanghai, China.Front Microbiol. 2018 Jul 31;9:1642. doi: 10.3389/fmicb.2018.01642. eCollection 2018. Front Microbiol. 2018. PMID: 30108555 Free PMC article.
-
Comparison of the Filtration Culture and Multiple Real-Time PCR Examination for Campylobacter spp. From Stool Specimens in Diarrheal Patients.Front Microbiol. 2018 Dec 5;9:2995. doi: 10.3389/fmicb.2018.02995. eCollection 2018. Front Microbiol. 2018. PMID: 30568645 Free PMC article.
References
-
- Euzeby JP. List of prokaryotic names with standing in nomenclature. Genus Campylobacter. http://www.bacterio.net/campylobacter.html.
-
- WHO. 2011. Campylobacter. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs255/en/.
-
- ESR. 2006. Notifiable and other diseases in New Zealand: annual report 2006. Institute of Environmental Science and Research Ltd, Porirua, New Zealand.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

