Mechanism of Antiactivation at the Pseudomonas sp. Strain ADP σN-Dependent PatzT Promoter
- PMID: 27208099
- PMCID: PMC4959210
- DOI: 10.1128/AEM.00906-16
Mechanism of Antiactivation at the Pseudomonas sp. Strain ADP σN-Dependent PatzT Promoter
Abstract
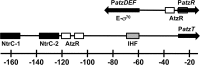
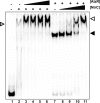
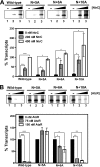
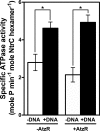
PatzT is an internal promoter of the atzRSTUVW operon that directs the synthesis of AtzT, AtzU, AtzV, and AtzW, components of an ABC-type cyanuric acid transport system. PatzT is σ(N) dependent, activated by the general nitrogen control regulator NtrC with the assistance of protein integration host factor (IHF), and repressed by the LysR-type transcriptional regulator (LTTR) AtzR. We have used a variety of in vivo and in vitro gene expression and protein-DNA interaction assays to assess the mechanisms underlying AtzR-dependent repression of PatzT Here, we show that repression only occurs when AtzR and NtrC interact simultaneously with the PatzT promoter region, indicating that AtzR acts as an antiactivator to antagonize activation by NtrC. Furthermore, repression requires precise rotational orientation of the AtzR and NtrC binding sites, strongly suggesting protein-protein interaction between the two proteins on the promoter region. Further exploration of the antiactivation mechanism showed that although AtzR-dependent repression occurs prior to open complex formation, AtzR does not alter the oligomerization state of NtrC or inhibit NtrC ATPase activity when bound to the PatzT promoter region. Taken together, these results strongly suggest that PatzT-bound AtzR interacts with NtrC to prevent the coupling of NtrC-mediated ATP hydrolysis with the remodeling of the interactions between E-σ(N) and PatzT that lead to open complex formation.
Importance: Here, we describe a unique mechanism by which the regulatory protein AtzR prevents the activation of the σ(N)-dependent promoter PatzT Promoters of this family are always positively regulated, but there are a few examples of overlapping negative regulation. The mechanism described here is highly unconventional and involves an interaction between the repressor and activator proteins to prevent the action of the repressor protein on the RNA polymerase-promoter complex.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures





Similar articles
-
Transcriptional organization and regulatory elements of a Pseudomonas sp. strain ADP operon encoding a LysR-type regulator and a putative solute transport system.J Bacteriol. 2012 Dec;194(23):6560-73. doi: 10.1128/JB.01348-12. Epub 2012 Oct 5. J Bacteriol. 2012. PMID: 23042989 Free PMC article.
-
Activation and repression of a sigmaN-dependent promoter naturally lacking upstream activation sequences.Mol Microbiol. 2009 Aug;73(3):419-33. doi: 10.1111/j.1365-2958.2009.06779.x. Epub 2009 Jun 29. Mol Microbiol. 2009. PMID: 19570137
-
Regulation of the Pseudomonas sp. strain ADP cyanuric acid degradation operon.J Bacteriol. 2005 Jan;187(1):155-67. doi: 10.1128/JB.187.1.155-167.2005. J Bacteriol. 2005. PMID: 15601699 Free PMC article.
-
Regulation of the atrazine-degradative genes in Pseudomonas sp. strain ADP.FEMS Microbiol Lett. 2010 Sep 1;310(1):1-8. doi: 10.1111/j.1574-6968.2010.01991.x. Epub 2010 Apr 15. FEMS Microbiol Lett. 2010. PMID: 20497226 Review.
-
Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators.Annu Rev Microbiol. 1997;51:341-73. doi: 10.1146/annurev.micro.51.1.341. Annu Rev Microbiol. 1997. PMID: 9343354 Review.
Cited by
-
A Fur family protein BosR is a novel RNA-binding protein that controls rpoS RNA stability in the Lyme disease pathogen.Nucleic Acids Res. 2024 May 22;52(9):5320-5335. doi: 10.1093/nar/gkae114. Nucleic Acids Res. 2024. PMID: 38366569 Free PMC article.
-
Fundamentals and Exceptions of the LysR-type Transcriptional Regulators.ACS Synth Biol. 2024 Oct 18;13(10):3069-3092. doi: 10.1021/acssynbio.4c00219. Epub 2024 Sep 22. ACS Synth Biol. 2024. PMID: 39306765 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

