Of the Nine Cytidine Deaminase-Like Genes in Arabidopsis, Eight Are Pseudogenes and Only One Is Required to Maintain Pyrimidine Homeostasis in Vivo
- PMID: 27208239
- PMCID: PMC4902590
- DOI: 10.1104/pp.15.02031
Of the Nine Cytidine Deaminase-Like Genes in Arabidopsis, Eight Are Pseudogenes and Only One Is Required to Maintain Pyrimidine Homeostasis in Vivo
Abstract
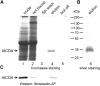
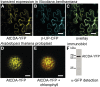
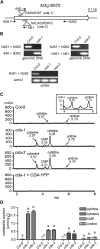
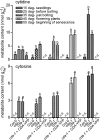
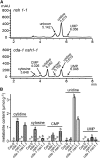
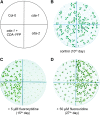
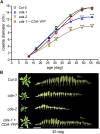
CYTIDINE DEAMINASE (CDA) catalyzes the deamination of cytidine to uridine and ammonia in the catabolic route of C nucleotides. The Arabidopsis (Arabidopsis thaliana) CDA gene family comprises nine members, one of which (AtCDA) was shown previously in vitro to encode an active CDA. A possible role in C-to-U RNA editing or in antiviral defense has been discussed for other members. A comprehensive bioinformatic analysis of plant CDA sequences, combined with biochemical functionality tests, strongly suggests that all Arabidopsis CDA family members except AtCDA are pseudogenes and that most plants only require a single CDA gene. Soybean (Glycine max) possesses three CDA genes, but only two encode functional enzymes and just one has very high catalytic efficiency. AtCDA and soybean CDAs are located in the cytosol. The functionality of AtCDA in vivo was demonstrated with loss-of-function mutants accumulating high amounts of cytidine but also CMP, cytosine, and some uridine in seeds. Cytidine hydrolysis in cda mutants is likely caused by NUCLEOSIDE HYDROLASE1 (NSH1) because cytosine accumulation is strongly reduced in a cda nsh1 double mutant. Altered responses of the cda mutants to fluorocytidine and fluorouridine indicate that a dual specific nucleoside kinase is involved in cytidine as well as uridine salvage. CDA mutants display a reduction in rosette size and have fewer leaves compared with the wild type, which is probably not caused by defective pyrimidine catabolism but by the accumulation of pyrimidine catabolism intermediates reaching toxic concentrations.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures
References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Betts L, Xiang S, Short SA, Wolfenden R, Carter CW Jr (1994) Cytidine deaminase: the 2.3 A crystal structure of an enzyme:transition-state analog complex. J Mol Biol 235: 635–656 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

