mRNA Decay of Most Arabidopsis miRNA Targets Requires Slicer Activity of AGO1
- PMID: 27208258
- PMCID: PMC4972266
- DOI: 10.1104/pp.16.00231
mRNA Decay of Most Arabidopsis miRNA Targets Requires Slicer Activity of AGO1
Abstract
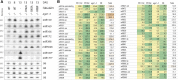
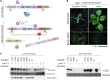
MicroRNAs (miRNAs) are key posttranscriptional regulators of gene expression in animals and plants. They guide RNA-induced silencing complexes to complementary target mRNA, thereby mediating mRNA degradation or translational repression. ARGONAUTE (AGO) proteins bind directly to miRNAs and may catalyze cleavage (slicing) of target mRNAs. In animals, miRNA target degradation via slicing occurs only exceptionally, and target mRNA decay is induced via AGO-dependent recruitment of deadenylase complexes. Conversely, plant miRNAs generally direct slicing of their targets, but it is unclear whether slicer-independent mechanisms of target mRNA decay also exist, and, if so, how much they contribute to miRNA-induced mRNA decay. Here, we compare phenotypes and transcript profiles of ago1 null and slicer-deficient mutants in Arabidopsis (Arabidopsis thaliana). We also construct conditional loss-of-function mutants of AGO1 to allow transcript profiling in true leaves. Although phenotypic differences between ago1 null and slicer-deficient mutants can be discerned, the results of both transcript profiling approaches indicate that slicer activity is required for mRNA repression of the vast majority of miRNA targets. A set of genes exhibiting up-regulation specifically in ago1 null, but not in ago1 slicer-deficient mutants was also identified, leaving open the possibility that AGO1 may have functions in gene regulation independent of small RNAs.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures



References
-
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 - PubMed
-
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. (2012) Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 19: 998–1004 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

